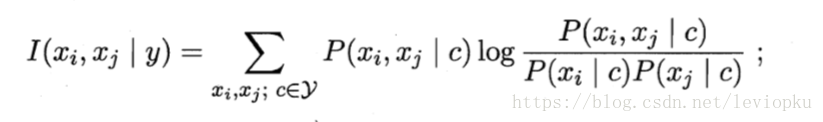概率机器学习的系列博文已经写到第四篇了,依然关注者乏。虽说来只是自己的学习记录,不过这样下去显得自己贡献值比较低下。于是今天换一种模式来写博文——结合代码实现。
欢迎各位指点交流~
预备知识:
1.朴素贝叶斯算法
2.Python代码阅读能力:基于python3.6实现,无须安装机器学习模块。
1. 半朴素贝叶斯的“半”
前面说到,朴素贝叶斯(NB)的‘朴素’就体现在它假设各属性之间没有相互依赖,可以简化贝叶斯公式中P(x|c)的计算。但事实上,属性直接完全没有依赖的情况是非常少的。如果简单粗暴用朴素贝叶斯建模,泛化能力和鲁棒性都难以达到令人满意。
这就可以引出主角半朴素贝叶斯了,它假定每个属性最多只依赖一个(或k个)其他属性。它考虑属性间的相互依赖,但假定依赖只有一个(ODE)或k个(kDE)其他属性。这就是半朴素贝叶斯的’半’所体现之处。所以有:
表示 的父属性
2. 常见的半朴素贝叶斯算法SPODE和TAN
SPODE算法:假设所有属性都依赖同一个属性,这个属性称为“超父”属性。
TAN算法:通过最大带权生成树算法确定属性之间的依赖关系,简单点说,就是每个属性找到跟自己最相关的属性,然后形成一个有向边(只能往一个方向)。
下面的图借自周志华老师西瓜书:

3. 详解TAN算法
关于TAN算法的实现步骤,网上有很严谨的说明。我尽量用好理解的话解释TAN的步骤,不太严谨切勿深究。步骤如下:
- 计算任意两个属性之间的条件互信息(CMI,即相互依赖程度)
以每个属性为节点( ),CMI为边( )形成一张图。找到这张图的最大带权生成树。即找到一个节点之间的连接规则,这个规则满足三个条件:
- 能够连接所有节点;
- 使用最少数目的边;
- 边长(CMI)总和最大
再把节点连接关系设置为有向,即从父节点指向子节点。在这里把最先出现的属性设置为根节点,再由根节点出发来确定边的方向。在代码中把第2步和第3步一起执行。
- 求
代码,训练数据和测试数据都在我的github上有:
https://github.com/leviome/TAN
import re as pattern
import sys
import math
if len(sys.argv) != 4:
train = open('lymph_train.arff')
test = open('lymph_test.arff')
mode = 't'
else:
train = open(sys.argv[1])
test = open(sys.argv[2])
mode = sys.argv[3]
# name list of different features : ['A1', 'A2', 'A3', 'A4', 'A5', 'A8', 'A14', 'A15']
feature_names = []
# value lists for different features : len(value)=len(type), each inner list includes all value for one feature
# [ [..], [..], [..], [..]', [..], [..], [..], [..] ]
feature_values = []
# value list for different labels : ['+', '-']
class_list = []
# list of training data (features)
train_data = []
# list of training data (labels)
train_data_label = []
# list of testing data (features)
test_data = []
# list of testing data (labels)
test_data_label = []
# calculate CMI between i feature and j feature, traverse all possible values and classes
# return CMI value between feature[I] and feature[J]
def calculate_conditional_mutual_information(I, J):
# list for all possible values within a certain feature
XI = feature_values[I]
XJ = feature_values[J]
CMI = 0.0
# get the list of combination of XI, XJ and all classes
XI_XJ_Class = list([])
for k in range(len(train_data)):
XI_XJ_Class.append([train_data[k][I], train_data[k][J], train_data_label[k]])
# get the list of [XI], [XJ], [XI,XJ]
for single_class in class_list:
# extract column with same class and certain feature
feature_is_XI = extract_feature_col(train_data_label, single_class, train_data, I, None)
feature_is_XJ = extract_feature_col(train_data_label, single_class, train_data, J, None)
feature_is_XIXJ = extract_feature_col(train_data_label, single_class, train_data, I, J)
for i in range(len(XI)):
# single value of XI
xi = XI[i]
for j in range(len(XJ)):
# single value of XJ
xj = XJ[j]
# calculate conditional possibility of xi, xj, or xi,xj , given class y
# need to match xi or xj value with all train data within same class
possibility_xi_given_y = laplace_estimate_possibility(feature_is_XI, [xi], len(feature_values[I]))
possibility_xj_given_y = laplace_estimate_possibility(feature_is_XJ, [xj], len(feature_values[J]))
possibility_xixj_given_y = laplace_estimate_possibility(feature_is_XIXJ, [xi, xj], len(feature_values[I]) * len(feature_values[J]))
possibility_xixjy = laplace_estimate_possibility(XI_XJ_Class, [xi, xj, single_class], len(feature_values[I]) * len(feature_values[J]) * len(class_list))
CMI = CMI + possibility_xixjy * math.log(possibility_xixj_given_y / (possibility_xi_given_y * possibility_xj_given_y), 2)
return CMI
# extract train_data (only the columns of index_of_feature) by checking whether train_data_label == goal_class_value
# return 1 column or 2 columns which has the same class
def extract_feature_col(class__train, goal_class_value, feature__train, index1_of_feature, index2_of_feature):
col_of_certain_feature = list([])
# traverse all train data
for i in range(len(class__train)):
if class__train[i] == goal_class_value:
tem = list([])
# row: i, column: I, in train data
tem.append(feature__train[i][index1_of_feature])
if index2_of_feature is not None:
tem.append(feature__train[i][index2_of_feature])
# make a list in a list: [ [XI or XJ or [XI,XJ], ... , [XI or XJ or [XI,XJ] ]
col_of_certain_feature.append(tem)
return col_of_certain_feature
# in the same class, each xi or xj value's ratio/possibility;
# return a possibility value
def laplace_estimate_possibility(feature_column_with_same_class, feature_value, amount_of_values_combination):
num = 0
for i in range(len(feature_column_with_same_class)):
if feature_column_with_same_class[i] == feature_value:
num += 1
# numerator + 1 to avoid 0; corresponds to denominator add the amount of all value combination
return float(num + 1) / (len(feature_column_with_same_class) + amount_of_values_combination)
# find a maximum search tree using the edges existed; finally, each node has and has only one parent
# return a matrix (graph) which represents the maximum search tree (row is parent, column is child)
def prims_algorithm(edges, graph):
all_candidates = set(range(0, len(feature_names)))
# To root the maximal weight spanning tree, pick the first attribute (index=0) in the input file as the root
parent_candidates = set()
parent_candidates.add(0)
child_candidates = set(range(1, len(feature_names)))
parent_child_list = list([])
# If there are ties in selecting maximum weight edges, use the following preference criteria:
# 1. Prefer edges emanating from attributes listed earlier in the input file.
while parent_candidates != all_candidates:
current_max = float('-inf')
parent = None
child = None
for i in parent_candidates:
for j in child_candidates:
# 2. If there are multiple maximal weight edges emanating from the first such attribute,
# prefer edges going to attributes listed earlier in the input file.
if edges[i][j] <= current_max:
pass
elif edges[i][j] > current_max:
parent = i
child = j
current_max = edges[i][j]
parent_child_list.append([parent, child])
parent_candidates.add(child)
child_candidates.remove(child)
# Finally, in parent_child_list, each child [i][1] will appear only once,no repetition, totally len(features)-1
# (exclusively 0, namely the root)
for i in range(len(parent_child_list)):
graph[parent_child_list[i][0]][parent_child_list[i][1]] = True
# print(parent_child_list)
# return conditional probability p(child_node_value | class_node_value, parent_node_value),
# given class_value and parent_value
def conditional_probability(child_node_value, class_node_value, parent_node_value, parent_feature_index, child_feature_index):
# extract the column of child_feature_index from the segment which has parent_node_value and class_node_value
child_column = list([])
for i in range(len(train_data)):
if train_data[i][parent_feature_index] == parent_node_value:
if train_data_label[i] == class_node_value:
child_column.append(train_data[i][child_feature_index])
# child_column has all values of feature[child_feature_index]
num = 0
for i in range(len(child_column)):
if child_column[i] == child_node_value:
num += 1
# numerator + 1 to avoid 0; corresponds to denominator add the amount of all value combination
return float(num + 1) / (len(child_column) + len(feature_values[child_feature_index]))
# return a dictionary includes all cases for all XI, XJ, class values (traverse 3 "for")
# key order is child_value_index + class_value_index + parent_value_index
def create_dictionary_of_conditional_probability(parent_feature_index, child_feature_index):
parent_feature = feature_values[parent_feature_index]
child_feature = feature_values[child_feature_index]
dictionary = {}
for i in range(len(parent_feature)):
for j in range(len(child_feature)):
for k in range(len(class_list)):
# p(child_value | class_value, parent_value)
key = str(j) + str(k) + str(i)
dictionary[key] = conditional_probability(child_feature[j], class_list[k], parent_feature[i], parent_feature_index, child_feature_index)
return dictionary
# return p( X | class_list[class_value_index] ), X is determined by a_row_test_data
def prior_probability(graph, dictionaries, a_row_test_data, class_value_index):
single_prior_probability = list([])
chain_rule_prior_probability = 1.0
for child in range(len(a_row_test_data)):
parent_value_index = None
child_value_index = None
# find current child_feature's parent_feature and their value_index
for parent in range(len(a_row_test_data)):
if graph[parent][child] == 1:
# find parent_value_index using parent_value in a_row_test_data
for parent_value_index in range(len(feature_values[parent])):
if feature_values[parent][parent_value_index] == a_row_test_data[parent]:
break
# find child_value_index using child_value in a_row_test_data
for child_value_index in range(len(feature_values[child])):
if feature_values[child][child_value_index] == a_row_test_data[child]:
break
break
if child == 0: # root
# find the child_value_index inside the root feature, using the a_row_test_data[child]
for child_value_index in range(len(feature_values[child])):
if feature_values[child][child_value_index] == a_row_test_data[child]:
break
# p( x0 | class_list[class_value_index])
single_prior_probability.append(dictionaries[child][str(child_value_index) + str(class_value_index)])
else: # other nodes
# p( xj | class_list[class_value_index], x_parent )
single_prior_probability.append(dictionaries[child][str(child_value_index) + str(class_value_index) + str(parent_value_index)])
for i in range(len(single_prior_probability)):
chain_rule_prior_probability = chain_rule_prior_probability * single_prior_probability[i]
return chain_rule_prior_probability
# binary classification problem
def TAN_predict(graph, dictionaries, a_row_test_data):
probability_list = list([]) # for all class values
sum = 0.0
# class_value_index is i
for i in range(len(class_list)):
# p(yi)
probability_of_yi = laplace_estimate_possibility(train_data_label, class_list[i], len(class_list))
# compute prior probability p( X | yi )
probability_of_feature_i = prior_probability(graph, dictionaries, a_row_test_data, i)
probability_list.append(probability_of_yi * probability_of_feature_i)
# the sum of all probability_list[i] is not 1 !!!
sum += probability_list[i]
max_probability = float('-inf')
y_predict_index = 0
for i in range(len(probability_list)):
# now, the sum of all probability_list[i] is 1
# now, probability_list[i] is posterior probability p( y_predict | test_data[i] )
probability_list[i] = probability_list[i] / sum
if probability_list[i] > max_probability:
max_probability = probability_list[i]
y_predict_index = i
return [class_list[y_predict_index], max_probability]
# compute edges' weights -> find maximum spanning tree (graph) ->
# according to the graph, construct a dictionary includes all combinations' conditional probability (dictionaries) ->
# classify test_data using Bayes Net chain rules (y_predict and probability_is_predict)
def TAN():
# weights' matrix ( k by k, k is the length of features)
edges = list([])
for i in range(len(feature_names)):
edges.append([])
for j in range(len(feature_names)):
edges[i].append(0)
# calculate_edges(weights)
for i in range(len(feature_names)):
for j in range(i + 1, len(feature_names)):
edges[i][j] = calculate_conditional_mutual_information(i, j)
edges[j][i] = edges[i][j]
# find maximum weight spanning tree (MST)
graph = list([])
for i in range(len(feature_names)):
graph.append([])
for j in range(len(feature_names)):
graph[i].append(0)
prims_algorithm(edges, graph)
# print(graph)
# output the maximum weight spanning tree with edge directions
# after prims_algorithm, graph matrix's each column should have and have only one True
# (each node have only one parent, except the root)
for j in range(len(graph)):
if j == 0: # the root
print('1: %s class' % feature_names[j])
else: # other nodes
for i in range(len(graph)):
if graph[i][j] is True:
# output child first and then parent
print('2: %s %s class' % (feature_names[j], feature_names[i]))
break
print('')
# construct a big dictionary to store all possible conditional probability according to "graph" structure
length = len(feature_names)
dictionaries = list([{}] * length)
for j in range(len(graph)): # child_feature_index
if j == 0: # the root
# root: p( x0 | all y )
dictionary = {}
for i in range(len(feature_values[j])): # root feature's all values
for k in range(len(class_list)):
# certain class_value
feature_is_xroot = extract_feature_col(train_data_label, class_list[k], train_data, j, None)
# get the ratio of certain feature_value in the column above
# p ( xroot_value | class_value)
dictionary[str(i) + str(k)] = laplace_estimate_possibility(feature_is_xroot, [feature_values[j][i]], len(feature_values[j]))
dictionaries[j] = dictionary
else: # other nodes
for i in range(len(graph)): # parent_feature_index
if graph[i][j] is True:
dictionaries[j] = create_dictionary_of_conditional_probability(i, j)
# print(dictionaries)
# output: (i) the predicted class, (ii) the actual class, (iii) and the posterior probability of the predicted class
correct = 0
result = list([])
for i in range(len(test_data)):
[y_predict, probability_is_predict] = TAN_predict(graph, dictionaries, test_data[i])
dummy = [y_predict, probability_is_predict]
result.append(dummy)
for i in range(len(result)):
if result[i][0] == test_data_label[i]:
correct += 1
print('3: %s %s %.12f' % (result[i][0], test_data_label[i], result[i][1]))
print('')
print(correct)
# comopute p(xj|class_value) -> p(X|class_value) -> p(X|class_value)*p(class_value) (nominator) ->
# sum all p(X|class_value)*p(class_value) for all class_value (denominator) ->
# p(class_value|X) (the probability of "prediction is class_value") ->
# find max probability for all class_value, namely prediction label
def naive_bayes():
correct = 0
probability_is_class_i = list([])
for i in range(len(feature_names)):
print(feature_names[i] + ' class')
print('')
# pre-set memory
for t in range(len(test_data)):
probability_is_class_i.append([])
# get p(Y|X) for each y of each test samples
for t in range(len(test_data)):
sample_data = test_data[t]
# get p(Y|X) for each y
for i in range(len(class_list)):
# p(X|class_value) = p(x1|class_value) * p(x2|class_value) * ... * p(xj|class_value)
probability_X_given_class_value = 1.0
for k in range(len(feature_names)):
temp_list = extract_feature_col(train_data_label, class_list[i], train_data, k, None)
probability_X_given_class_value *= laplace_estimate_possibility(temp_list, [sample_data[k]], len(feature_values[k]))
# p(class_value) * p(X | class_value)
probability_class_value = laplace_estimate_possibility(train_data_label, class_list[i], len(class_list))
probability_is_class_i[t].append(probability_class_value * probability_X_given_class_value)
# find max probability as prediction and match it with ture label
for t in range(len(test_data)):
denominator = 0.0
max_probability = 0.0
index_of_class_value_prediction = 0
# get bayes rule's denominator
for i in range(len(class_list)):
denominator += float(probability_is_class_i[t][i])
# p(class_list[i] | X)
for i in range(len(class_list)):
probability_is_class_i[t][i] = probability_is_class_i[t][i] / denominator
if probability_is_class_i[t][i] > max_probability:
max_probability = probability_is_class_i[t][i]
index_of_class_value_prediction = i
# match prediction with true label
if class_list[index_of_class_value_prediction] == test_data_label[t]:
correct += 1
print('%s %s %.12f' % (class_list[index_of_class_value_prediction], test_data_label[t], max_probability))
print('')
print(correct)
begin_data = False
for line in train:
if pattern.findall('@data', line) != []:
begin_data = True
elif pattern.findall('@attribute', line) != []:
line = line.lstrip(' ')
line = line.rstrip('\n')
line = line.rstrip('\r')
line = line.rstrip(' ')
line = line.split(None, 2)
line[1] = line[1].replace(' ', '')
line[1] = line[1].replace('\'', '')
line[2] = line[2].replace(' ', '')
line[2] = line[2].replace('\'', '')
line[2] = line[2].strip('{')
line[2] = line[2].strip('}')
line[2] = line[2].split(',')
if line[1] != 'class':
feature_names.append(line[1])
feature_values.append(line[2])
else:
class_list = line[2]
elif begin_data is True:
line = line.strip('\n')
line = line.strip('\r')
line = line.replace(' ', '')
line = line.replace('\'', '')
line = line.split(',')
temp = []
for i in range(0, len(line) - 1):
temp.append(line[i])
train_data.append(temp)
train_data_label.append(line[len(line) - 1])
else:
pass
begin_data = False
for line in test:
if pattern.findall('@data', line) != []:
begin_data = True
elif begin_data is True:
line = line.strip('\n')
line = line.strip('\r')
line = line.replace(' ', '')
line = line.replace('\'', '')
line = line.split(',')
temp = []
for i in range(0, len(line) - 1):
temp.append(line[i])
test_data.append(temp)
test_data_label.append(line[len(line) - 1])
else:
pass
if mode == 'n':
naive_bayes()
elif mode == 't':
TAN()
else:
pass