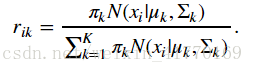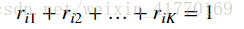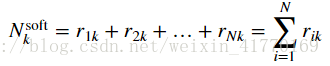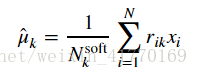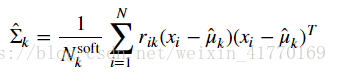1 k-means和MoG都容易出现局部最优解
2、公式:
3、自己实现EM算法
(1)主函数:
def EM(data, init_means, init_covariances, init_weights, maxiter=1000, thresh=1e-4):
# Make copies of initial parameters, which we will update during each iteration
means = init_means[:]
covariances = init_covariances[:]
weights = init_weights[:]
# Infer dimensions of dataset and the number of clusters
num_data = len(data)
num_dim = len(data[0])
num_clusters = len(means)
# Initialize some useful variables
resp = np.zeros((num_data, num_clusters))
ll = loglikelihood(data, weights, means, covariances)
ll_trace = [ll]
for it in range(maxiter):
if it % 5 == 0:
print("Iteration %s" % it)
# E-step: compute responsibilities
resp = compute_responsibilities(data, weights, means, covariances)
# M-step
# Compute the total responsibility assigned to each cluster, which will be useful when
# implementing M-steps below. In the lectures this is called N^{soft}
counts = compute_soft_counts(resp)
# Update the weight for cluster k using the M-step update rule for the cluster weight, \hat{\pi}_k.
# YOUR CODE HERE
weights = compute_weights(counts)
# Update means for cluster k using the M-step update rule for the mean variables.
# This will assign the variable means[k] to be our estimate for \hat{\mu}_k.
# YOUR CODE HERE
means = compute_means(data, resp, counts)
# Update covariances for cluster k using the M-step update rule for covariance variables.
# This will assign the variable covariances[k] to be the estimate for \hat{\Sigma}_k.
# YOUR CODE HERE
covariances = compute_covariances(data, resp, counts, means)
# Compute the loglikelihood at this iteration
# YOUR CODE HERE
ll_latest = loglikelihood(data, weights, means, covariances)
ll_trace.append(ll_latest)
# Check for convergence in log-likelihood and store
if (ll_latest - ll) < thresh and ll_latest > -np.inf:
break
ll = ll_latest
if it % 5 != 0:
print("Iteration %s" % it)
out = {'weights': weights, 'means': means, 'covs': covariances, 'loglik': ll_trace, 'resp': resp}
return out
(2)子函数:
def log_sum_exp(Z):
""" Compute log(\sum_i exp(Z_i)) for some array Z."""
return np.max(Z) + np.log(np.sum(np.exp(Z - np.max(Z))))
def loglikelihood(data, weights, means, covs):
""" Compute the loglikelihood of the data for a Gaussian mixture model with the given parameters. """
num_clusters = len(means)
num_dim = len(data[0])
ll = 0
for d in data:
Z = np.zeros(num_clusters)
for k in range(num_clusters):
# Compute (x-mu)^T * Sigma^{-1} * (x-mu)
delta = np.array(d) - means[k]
exponent_term = np.dot(delta.T, np.dot(np.linalg.inv(covs[k]), delta))
# Compute loglikelihood contribution for this data point and this cluster
Z[k] += np.log(weights[k])
Z[k] -= 1/2. * (num_dim * np.log(2*np.pi) + np.log(np.linalg.det(covs[k])) + exponent_term)
# Increment loglikelihood contribution of this data point across all clusters
ll += log_sum_exp(Z)
return ll
def compute_responsibilities(data, weights, means, covariances):
'''E-step: compute responsibilities, given the current parameters'''
num_data = len(data)
num_clusters = len(means)
resp = np.zeros((num_data, num_clusters))
# Update resp matrix so that resp[i,k] is the responsibility of cluster k for data point i.
# Hint: To compute likelihood of seeing data point i given cluster k, use multivariate_normal.pdf.
for i in range(num_data):
for k in range(num_clusters):
# YOUR CODE HERE
gdi = weights[k] * multivariate_normal.pdf(data[i], means[k], covariances[k])
gdall = sum([weights[kidx] * multivariate_normal.pdf(data[i], means[kidx], covariances[kidx]) for kidx in range(num_clusters)])
resp[i, k] = gdi/gdall
# Add up responsibilities over each data point and normalize
row_sums = resp.sum(axis=1)[:, np.newaxis]
resp = resp / row_sums
return resp
def compute_soft_counts(resp):
# Compute the total responsibility assigned to each cluster, which will be useful when
# implementing M-steps below. In the lectures this is called N^{soft}
counts = np.sum(resp, axis=0)
return counts
def compute_weights(counts):
num_clusters = len(counts)
weights = [0.] * num_clusters
for k in range(num_clusters):
# Update the weight for cluster k using the M-step update rule for the cluster weight, \hat{\pi}_k.
# HINT: compute # of data points by summing soft counts.
# YOUR CODE HERE
weights[k] = counts[k]/sum([counts[i] for i in range(num_clusters)])
return weights
def compute_means(data, resp, counts):
num_clusters = len(counts)
num_data = len(data)
means = [np.zeros(len(data[0]))] * num_clusters
for k in range(num_clusters):
# Update means for cluster k using the M-step update rule for the mean variables.
# This will assign the variable means[k] to be our estimate for \hat{\mu}_k.
weighted_sum = 0.
for i in range(num_data):
# YOUR CODE HERE
weighted_sum += resp[i,k]*data[i]
# YOUR CODE HERE
means[k] = weighted_sum/counts[k]
return means
def compute_covariances(data, resp, counts, means):
num_clusters = len(counts)
num_dim = len(data[0])
num_data = len(data)
covariances = [np.zeros((num_dim,num_dim))] * num_clusters
for k in range(num_clusters):
# Update covariances for cluster k using the M-step update rule for covariance variables.
# This will assign the variable covariances[k] to be the estimate for \hat{\Sigma}_k.
weighted_sum = np.zeros((num_dim, num_dim))
for i in range(num_data):
# YOUR CODE HERE (Hint: Use np.outer on the data[i] and this cluster's mean)
weighted_sum += resp[i,k]*np.outer(data[i]-means[k],data[i]-means[k])
# YOUR CODE HERE
covariances[k] = weighted_sum/counts[k]
return covariances
3、训练模型得到高斯模型参数
images = gl.SFrame('images.sf')
gl.canvas.set_target('ipynb')
import array
images['rgb'] = images.pack_columns(['red', 'green', 'blue'])['X4']
images.show()
np.random.seed(1) # Initalize parameters init_means = [images['rgb'][x] for x in np.random.choice(len(images), 4, replace=False)] cov = np.diag([images['red'].var(), images['green'].var(), images['blue'].var()]) init_covariances = [cov, cov, cov, cov] init_weights = [1/4., 1/4., 1/4., 1/4.] # Convert rgb data to numpy arrays img_data = [np.array(i) for i in images['rgb']] # Run our EM algorithm on the image data using the above initializations. # This should converge in about 125 iterations out = EM(img_data, init_means, init_covariances, init_weights)
ll = out['loglik']
plt.plot(range(len(ll)),ll,linewidth=4)
plt.xlabel('Iteration')
plt.ylabel('Log-likelihood')
plt.rcParams.update({'font.size':16})
plt.tight_layout()

此时得到高斯模型参数
4、测试图像类别
weights = out['weights']
means = out['means']
covariances = out['covs']
rgb = images['rgb']
N = len(images) # number of images
K = len(means) # number of clusters
assignments = [0]*N
probs = [0]*N
for i in range(N):
# Compute the score of data point i under each Gaussian component:
p = np.zeros(K)
for k in range(K):
p[k] = weights[k]*multivariate_normal.pdf(rgb[i], mean=means[k], cov=covariances[k])
# Compute assignments of each data point to a given cluster based on the above scores:
assignments[i] = np.argmax(p)
# For data point i, store the corresponding score under this cluster assignment:
probs[i] = np.max(p)
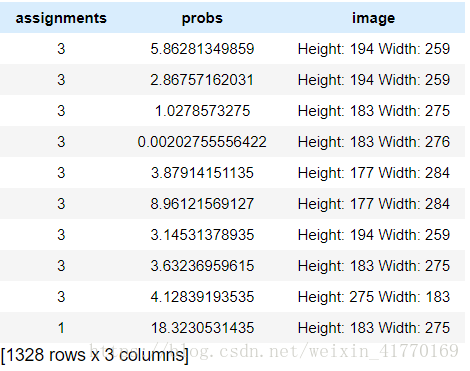
assignments = gl.SFrame({'assignments':assignments, 'probs':probs, 'image': images['image']})