【代码复现】SCGC__Simple Contrastive Graph Clustering
文章目录
1. 介绍
本文复现的代码为论文----Simple Contrastive Graph Clustering。
- 对比学习因其良好的性能而在深度图聚类中引起了广泛的关注。
- 然而,复杂的数据扩充和耗时的图卷积操作削弱了这些方法的效率。
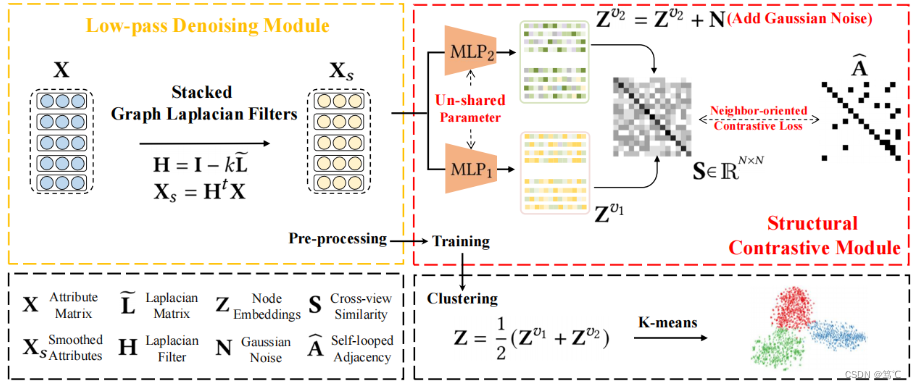
为了解决这一问题,作者提出了一个简单的对比图聚类(SCGC)算法,从网络架构、数据增强和目标函数的角度来改进现有的方法。在架构方面,网络包括两个主要部分,即预处理和网络骨干。
- 一个简单的低通去噪操作将邻居信息聚合作为一个独立的预处理,并且只包含两个多层感知器(MLPs)作为主干。
- 对于数据增强,模型没有在图上引入复杂的操作,而是通过设计参数非共享的暹罗编码器和直接干扰节点嵌入来构造同一顶点的两个增强视图。
- 最后,在目标函数方面,为了进一步提高聚类性能,设计了一种新的跨视图结构一致性目标函数,以提高学习网络的判别能力。
2. 前言
3. 复现代码
3.1 项目框架

3.2 代码文件
3.2.1 main.py
import tqdm
from torch.optim import Adam, SGD
from model import *
from utils import *
import opt
def train(X, y, A, a):
opt.args.acc, opt.args.nmi, opt.args.ari, opt.args.f1 = 0, 0, 0, 0
A_sl = a * A + np.eye(A.shape[0])
if opt.args.is_pass != 0:
if opt.args.is_pass == 1:
adjs = get_adjs(A)
for a in adjs:
X = a.dot(X)
else:
adjs = get_laps(A)
for a in adjs:
X = a.dot(X)
enc_dims = [opt.args.n_input] + opt.args.enc_dims
dec_dims = opt.args.dec_dims + [opt.args.n_input]
model = OUR(opt.args.layers, enc_dims, dec_dims).to(opt.args.device)
X = numpy_to_torch(X).to(opt.args.device)
A_sl = numpy_to_torch(A_sl).to(opt.args.device)
centers = model_init(model, X, y)
# initialize cluster centers
model.cluster_centers.data = torch.tensor(centers).to(opt.args.device)
optimizer = Adam(model.parameters(), lr=opt.args.lr)
for epoch in range(opt.args.epoch):
# input & output
Z1, Z2, Z, Q, X_ = model(X)
P = target_distribution(Q)
loss_cv = cross_view_loss(Z1, Z2, A_sl)
loss_kl = distribution_loss(Q, P)
loss_rec = reconstruction_loss(X, X_)
# print(loss_cv, loss_kl)
loss = loss_cv + 10 * loss_kl + loss_rec
# optimization
optimizer.zero_grad()
loss.backward(retain_graph=True)
optimizer.step()
# clustering & evaluation
acc, nmi, ari, f1, _ = clustering(Z, y)
if acc > opt.args.acc:
opt.args.acc = acc
opt.args.nmi = nmi
opt.args.ari = ari
opt.args.f1 = f1
print(epoch, "ACC: {:.4f},".format(acc), "NMI: {:.4f},".format(nmi), "ARI: {:.4f},".format(ari), "F1: {:.4f}".format(f1))
return opt.args.acc, opt.args.nmi, opt.args.ari, opt.args.f1
if __name__ == '__main__':
# initialize
setup()
# load data
X, y, A = load_graph_data(opt.args.name)
acc, nmi, ari, f1 = train(X, y, A, 1.0)
print("ACC: {:.4f},".format(acc), "NMI: {:.4f},".format(nmi), "ARI: {:.4f},".format(ari), "F1: {:.4f}".format(f1))
3.2.2 model.py
import torch.nn as nn
import torch.nn.functional as F
import torch
from utils import *
class LinTrans(nn.Module):
def __init__(self, layers, dims):
super(LinTrans, self).__init__()
self.layers = nn.ModuleList()
for i in range(layers):
self.layers.append(nn.Linear(dims[i], dims[i+1]))
self.act = nn.Sigmoid()
# self.act = nn.LeakyReLU()
def scale(self, z):
zmax = z.max(dim=1, keepdim=True)[0]
zmin = z.min(dim=1, keepdim=True)[0]
z_std = (z - zmin) / (zmax - zmin)
z_scaled = z_std
return z_scaled
def forward(self, x):
num_layer = len(self.layers)
out = x
for i in range(num_layer - 1):
out = self.act(self.layers[i](out))
out = self.layers[num_layer - 1](out)
# out = self.scale(out)
out = F.normalize(out)
return out
class OUR(nn.Module):
def __init__(self, lt_layers, dims):
super(OUR, self).__init__()
self.lt1 = LinTrans(lt_layers, dims)
self.lt2 = LinTrans(lt_layers, dims)
def forward(self, X):
Z1, Z2 = self.lt1(X), self.lt2(X)
return Z1, Z2
3.2.3 utils.py
import numpy as np
import scipy.sparse as sp
import matplotlib.pyplot as plt
import sklearn.preprocessing as preprocess
import torch
import random
import opt
import numpy as np
from sklearn import metrics
from munkres import Munkres
import torch.nn.functional as F
from sklearn.cluster import KMeans
from sklearn.decomposition import PCA
from sklearn.metrics import adjusted_rand_score as ari_score
from sklearn.metrics.cluster import normalized_mutual_info_score as nmi_score
def setup():
"""
setup
- name: the name of dataset
- device: CPU / GPU
- seed: random seed
- n_clusters: num of cluster
- n_input: dimension of feature
- alpha_value: alpha value for graph diffusion
- lambda_value: lambda value for clustering guidance
- gamma_value: gamma value for propagation regularization
- lr: learning rate
Return: None
"""
# print("---------------setting---------------")
setup_seed(opt.args.seed)
if opt.args.name == 'amap':
print('amap...............')
opt.args.n_clusters = 8
opt.args.t = 5
opt.args.lr = 1e-5
elif opt.args.name == 'cite':
print('cite...............')
opt.args.n_clusters = 6
opt.args.t = 2
opt.args.lr = 5e-5
elif opt.args.name == 'cora':
print('cora...............')
opt.args.n_clusters = 7
opt.args.t = 2
opt.args.lr = 1e-3
elif opt.args.name == 'corafull':
print('corafull...............')
opt.args.n_clusters = 70
opt.args.t = 2
opt.args.lr = 1e-4
elif opt.args.name == 'bat':
# opt.args.n_input = 50
print('bat...............')
opt.args.n_clusters = 4
opt.args.t = 3
opt.args.lr = 1e-3
elif opt.args.name == 'eat':
print('eat...............')
opt.args.n_clusters = 4
opt.args.t = 5
opt.args.lr = 1e-3
elif opt.args.name == 'uat':
print('uat...............')
opt.args.n_clusters = 4
opt.args.t = 3
opt.args.lr = 1e-3
else:
print("error!")
exit(0)
opt.args.device = torch.device("cuda:1" if opt.args.cuda else "cpu")
# opt.args.device = torch.device("cpu")
# print("dataset : {}".format(opt.args.name))
# print("device : {}".format(opt.args.device))
# print("random seed : {}".format(opt.args.seed))
# print("clusters : {}".format(opt.args.n_clusters))
# print("n_PCA : {}".format(opt.args.n_input))
# print("learning rate : {:.0e}".format(opt.args.lr))
def setup_seed(seed):
"""
setup random seed to fix the result
Args:
seed: random seed
Returns: None
"""
torch.manual_seed(seed)
torch.cuda.manual_seed(seed)
torch.cuda.manual_seed_all(seed)
np.random.seed(seed)
random.seed(seed)
torch.manual_seed(seed)
torch.backends.cudnn.benchmark = False
torch.backends.cudnn.deterministic = True
def numpy_to_torch(a, sparse=False):
"""
numpy array to torch tensor
:param a: the numpy array
:param sparse: is sparse tensor or not
:return: torch tensor
"""
if sparse:
a = torch.sparse.Tensor(a)
a = a.to_sparse()
else:
a = torch.FloatTensor(a)
return a
def torch_to_numpy(t):
"""
torch tensor to numpy array
:param t: the torch tensor
:return: numpy array
"""
return t.numpy()
def load_graph_data(dataset_name, show_details=False):
"""
load graph data
:param dataset_name: the name of the dataset
:param show_details: if show the details of dataset
- dataset name
- features' shape
- labels' shape
- adj shape
- edge num
- category num
- category distribution
:return: the features, labels and adj
"""
load_path = "../dataset/" + dataset_name + "/" + dataset_name
feat = np.load(load_path+"_feat.npy", allow_pickle=True)
label = np.load(load_path+"_label.npy", allow_pickle=True)
adj = np.load(load_path+"_adj.npy", allow_pickle=True)
if show_details:
print("++++++++++++++++++++++++++++++")
print("---details of graph dataset---")
print("++++++++++++++++++++++++++++++")
print("dataset name: ", dataset_name)
print("feature shape: ", feat.shape)
print("label shape: ", label.shape)
print("adj shape: ", adj.shape)
print("undirected edge num: ", int(np.nonzero(adj)[0].shape[0]/2))
print("category num: ", max(label)-min(label)+1)
print("category distribution: ")
for i in range(max(label)+1):
print("label", i, end=":")
print(len(label[np.where(label == i)]))
print("++++++++++++++++++++++++++++++")
# X pre-processing
# pca = PCA(n_components=opt.args.n_input)
# feat = pca.fit_transform(feat)
opt.args.n_input = feat.shape[1]
return feat, label, adj
def gaussian_noised_feature(X):
"""
add gaussian noise to the attribute matrix X
Args:
X: the attribute matrix
Returns: the noised attribute matrix Y
"""
N = torch.Tensor(np.random.normal(0, 0.01, X.shape)).to(opt.args.device)
Y = X + N
return Y
def gaussian_noised_feature_(X):
"""
add gaussian noise to the attribute matrix X
Args:
X: the attribute matrix
Returns: the noised attribute matrix Y
"""
N = torch.Tensor(np.random.normal(1, 0.01, X.shape)).to(opt.args.device)
Y = X * N
return Y
def normalize_adj(adj, self_loop=True, symmetry=False):
"""
normalize the adj matrix
:param adj: input adj matrix
:param self_loop: if add the self loop or not
:param symmetry: symmetry normalize or not
:return: the normalized adj matrix
"""
ident = np.eye(adj.shape[0])
# add the self_loop
if self_loop:
adj_tmp = adj + ident
else:
adj_tmp = adj
# calculate degree matrix and it's inverse matrix
row_sum = adj_tmp.sum(1)
L = np.diag(row_sum) - adj_tmp
if symmetry:
d1 = np.diag(np.power(row_sum, -0.5))
norm_L = np.matmul(np.matmul(d1, L), d1) # symmetry normalize: D^{-0.5} A D^{-0.5}
else:
d2 = np.diag(np.power(row_sum, -1))
norm_L = np.matmul(d2, L) # non-symmetry normalize: D^{-1} A
return norm_L
def get_adjs(adj, norm = True):
ident = 1 * np.eye(adj.shape[0])
norm_L = normalize_adj(adj, True, norm)
reg = [1] * (opt.args.t)
# reg = [1] * (2)
print('t...............', len(reg))
adjs = []
for i in range(len(reg)):
adjs.append(ident-(reg[i] * norm_L))
# for i in range(len(reg)):
# adjs.append(norm_L)
return adjs
# Calculating loss-----------------------------------------------------------------start
def distance(x, y):
return torch.sum(torch.square(x - y))
def similarity_loss(edges, Z):
num_edges = len(edges)
loss_sim = [0.0]
loss_sim = torch.FloatTensor(loss_sim).to(opt.args.device)
for i in range(0, num_edges):
loss_sim += distance(Z[edges[i][0]], Z[edges[i][1]])
return loss_sim / num_edges
def cross_correlation(X, Y):
return torch.mm(X, Y.t())
def cross_view_loss(X, Y, A):
# cross-view similarity matrix
S = cross_correlation(X, Y)
# loss of cross view
L_cv = (A-S).pow(2).mean()
return L_cv
def aug_loss(X, Xl, A, Al):
return - (A-Al).pow(2).mean() - (X-Xl).pow(2).mean()
# Calculating loss-----------------------------------------------------------------end
# Clustering and Evaluation--------------------------------------------------------start
def clustering(Z, y):
"""
clustering based on embedding
Args:
Z: the input embedding
y: the ground truth
Returns: acc, nmi, ari, f1, clustering centers
"""
model = KMeans(n_clusters=opt.args.n_clusters, n_init=20)
cluster_id = model.fit_predict(Z.data.cpu().numpy())
acc, nmi, ari, f1 = eva(y, cluster_id, show_details=opt.args.show_training_details)
return acc, nmi, ari, f1, model.cluster_centers_
def cluster_acc(y_true, y_pred):
"""
calculate clustering acc and f1-score
Args:
y_true: the ground truth
y_pred: the clustering id
Returns: acc and f1-score
"""
y_true = y_true - np.min(y_true)
l1 = list(set(y_true))
num_class1 = len(l1)
l2 = list(set(y_pred))
num_class2 = len(l2)
ind = 0
if num_class1 != num_class2:
for i in l1:
if i in l2:
pass
else:
y_pred[ind] = i
ind += 1
l2 = list(set(y_pred))
numclass2 = len(l2)
if num_class1 != numclass2:
print('error')
return
cost = np.zeros((num_class1, numclass2), dtype=int)
for i, c1 in enumerate(l1):
mps = [i1 for i1, e1 in enumerate(y_true) if e1 == c1]
for j, c2 in enumerate(l2):
mps_d = [i1 for i1 in mps if y_pred[i1] == c2]
cost[i][j] = len(mps_d)
m = Munkres()
cost = cost.__neg__().tolist()
indexes = m.compute(cost)
new_predict = np.zeros(len(y_pred))
for i, c in enumerate(l1):
c2 = l2[indexes[i][1]]
ai = [ind for ind, elm in enumerate(y_pred) if elm == c2]
new_predict[ai] = c
acc = metrics.accuracy_score(y_true, new_predict)
f1_macro = metrics.f1_score(y_true, new_predict, average='macro')
return acc, f1_macro
def eva(y_true, y_pred, show_details=False):
"""
evaluate the clustering performance
Args:
y_true: the ground truth
y_pred: the predicted label
show_details: if print the details
Returns: None
"""
acc, f1 = cluster_acc(y_true, y_pred)
nmi = nmi_score(y_true, y_pred, average_method='arithmetic')
ari = ari_score(y_true, y_pred)
if show_details:
print(':acc {:.4f}'.format(acc), ', nmi {:.4f}'.format(nmi), ', ari {:.4f}'.format(ari),
', f1 {:.4f}'.format(f1))
return acc, nmi, ari, f1
# Clustering and Evaluation--------------------------------------------------------end
3.2.4 opt.py
import argparse
parser = argparse.ArgumentParser(description='OUR', formatter_class=argparse.ArgumentDefaultsHelpFormatter)
# setting
parser.add_argument('--name', type=str, default="cite")
parser.add_argument('--cuda', type=bool, default=True)
parser.add_argument('--seed', type=int, default=42)
parser.add_argument('--lr', type=float, default=1e-4)
parser.add_argument('--layers', type=int, default=1)
parser.add_argument('--dims', type=int, default=[500], help='Number of units in hidden layer 1.')
parser.add_argument('--epoch', type=int, default=400)
parser.add_argument('--show_training_details', type=bool, default=False)
# clustering performance: acc, nmi, ari, f1
parser.add_argument('--acc', type=float, default=0)
parser.add_argument('--nmi', type=float, default=0)
parser.add_argument('--ari', type=float, default=0)
parser.add_argument('--f1', type=float, default=0)
args = parser.parse_args()
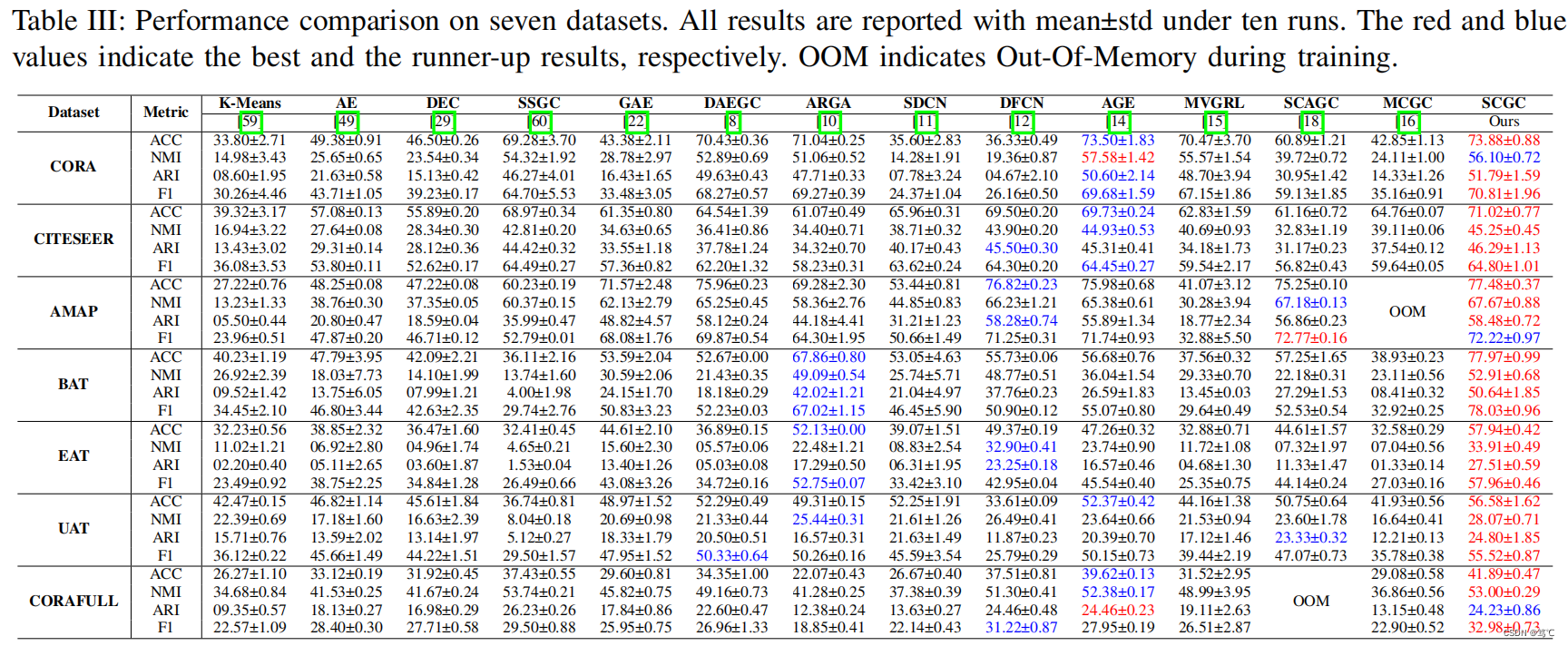
3.3 实验结果
4. 参考
【1】https://blog.csdn.net/qq_51392112/article/details/128943812
【2】https://blog.csdn.net/qq_51392112/article/details/129429108