决策树学习算法包含特征选择、决策树的生成与剪枝过程。决策树的学习算法一般是递归地选择最优特征,并用最优特征对数据集进行分割。由于决策树表示条件概率分布,所以高度不同的决策树对应不同复杂度的概率模型。最优决策树的生成是个NP问题,能实现的生成算法都是局部最优的,剪枝则是既定决策树下的全局最优。
A特征选择:
如何判断一个特征的分类能力呢?有以下两种方法:1、信息增益;2、信息增益比。
信息增益:
对于一个可能有n种取值的随机变量: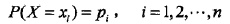 ,其熵为
,其熵为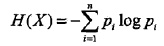 ,且
,且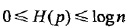 。
。
设有随机变量(X,Y),其联合分布为: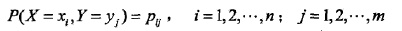 ,条件熵H(Y|X)表示在已知随机变量X的条件下随机变量Y的不确定性,定义为在X给定的条件下,Y的条件概率分布对X的数学期望:
,条件熵H(Y|X)表示在已知随机变量X的条件下随机变量Y的不确定性,定义为在X给定的条件下,Y的条件概率分布对X的数学期望: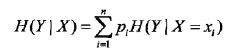 ,
,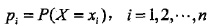 。当上述定义式中的概率由数据估计(比如上一章提到的极大似然估计)得到时,所对应的熵和条件熵分别称为经验熵和经验条件熵。则有了相关定义:
。当上述定义式中的概率由数据估计(比如上一章提到的极大似然估计)得到时,所对应的熵和条件熵分别称为经验熵和经验条件熵。则有了相关定义:

决策树学习中的信息增益等价于训练数据集中类与特征的互信息。具体计算流程如下:



信息增益比:
信息增益的值是相对于训练数据集而言的,当H(D)大的时候,信息增益值往往会偏大,这样对H(D)小的特征不公平。改进的方法是信息增益比:

ID3算法:
从根节点开始,计算所有可能的特征的信息增益,选择信息增益最大的特征作为当前节点的特征,由特征的不同取值建立空白子节点,对空白子节点递归调用此方法,直到所有特征的信息增益小于阀值或者没有特征可选为止。
C4.5算法:
C4.5算法与ID3相似,但是在选择的时候使用的是信息增益比,形式化地描述如下:

C剪枝:
决策树很容易发生过拟合,过拟合的原因在于学习的时候过多地考虑如何提高对训练数据的正确分类,从而构建出过于复杂的决策树。解决这个问题的办法就是简化已生成的决策树,也就是剪枝。决策树的剪枝往往通过极小化决策树整体的损失函数或代价函数来实现。设决策树T的叶节点有|T|个,t是某个叶节点,t有Nt个样本点,其中归入k类的样本点有Ntk个,Ht(T)为叶节点t上的经验熵,α≥0为参数,则损失函数可以定义为:
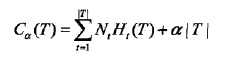
其中经验熵Ht(T)为:
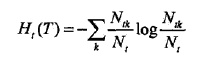
表示叶节点t所代表的类别的不确定性。损失函数对它求和表示所有被导向该叶节点的样本点所带来的不确定的和的和。我没有多打一个“的和”,第二个是针对叶节点t说的。
在损失函数中,将右边第一项记作:
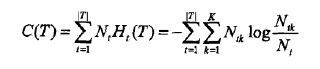
则损失函数可以简单记作:

C(T)表示模型对训练数据的预测误差,即模型与训练数据的拟合程度,|T|表示模型复杂度,参数α≥0控制两者之间的影响,α越大,模型越简单,α=0表示不考虑复杂度。
剪枝,就是当α确定时,选择损失函数最小的模型。子树越大C(T)越小,但是α|T|越大,损失函数反映的是两者的平衡。决策树的生成过程只考虑了信息增益或信息增益比,只考虑更好地拟合训练数据,而剪枝过程则考虑了减小复杂度。前者是局部学习,后者是整体学习。
3、CART算法
分类与回归树(CART)模型同样由特征选取、树的生成和剪枝组成,既可以用于分类也可以用于回归。CART假设决策树是二叉树,内部节点特征的取值为是和否,对应一个实例的特征是否是这样的。决策树递归地二分每个特征,将输入空间划分为有限个单元。
CART生成
递归地构建二叉决策树的过程。对回归树用平方误差最小化准则,对分类树用基尼指数最小化准则,进行特征选取,生成二叉树。
1、回归树
回归树与分类树在数据集上的不同就是数据集的输出部分不是类别,而是连续变量。假设输入空间已经被分为M个单元,分别对应输出值cm,于是回归树模型可以表示为:
回归树的预测误差:
那么输出值就是使上面误差最小的值,也就是均值:
难点在于怎么划分,一种启发式的方法(其实就是暴力搜索吧):
遍历所有输入变量,选择第j个变量和它的值s作为切分变量和切分点,将空间分为两个区域:
然后计算两个区域的平方误差,求和,极小化这个和,具体的,就是:
当j最优化的时候,就可以将切分点最优化:
递归调用此过程,这种回归树通常称为最小二乘回归树。
2、分类树
与回归树算法流程类似,只不过选择的是最优切分特征和最优切分点,并采用基尼指数衡量。基尼指数定义:
对于给定数据集D,其基尼指数是:
Ck是属于第k类的样本子集,K是类的个数。Gini(D)反应的是D的不确定性(与熵类似),分区的目标就是降低不确定性。
D根据特征A是否取某一个可能值a而分为D1和D2两部分:
则在特征A的条件下,D的基尼指数是:
有了上述知识储备,可以给出CART生成算法的伪码:
设节点的当前数据集为D,对D中每一个特征A,对齐每个值a根据D中样本点是否满足A==a分为两部分,计算基尼指数。对所有基尼指数选择最小的,对应的特征和切分点作为最优特征和最优切分点,生成两个子节点,将对应的两个分区分配过去,然后对两个子节点递归。
CART剪枝
在上面介绍的损失函数中,当α固定时,一定存在使得损失函数最小的子树,记为复杂度=Tα,α偏大Tα就偏小。设对α递增的序列,对应的最优子树序列为Tn,子树序列第一棵包含第二棵,依次类推。
从T0开始剪枝,对它内部的任意节点t,只有t这一个节点的子树的损失函数是:,以t为根节点的子树的损失函数是:
当α充分小,肯定有,这个不等式的意思是复杂模型在复杂度影响力小的情况下损失函数更小。当α增大到某一点,这个不等式的符号会反过来。只要,损失函数值就相同,但是t更小啊,所以t更可取,于是把Tt剪枝掉。为此,对每一个t,计算,表示损失函数的减少程度,从T中剪枝掉g(t)最小的Tt,取新的α=g(t),直到根节点。这样就得到了一个子树序列,对此序列,应用独立的验证数据集交叉验证,选取最优子树,剪枝完毕。
4、总结
决策树算法是机器学习中一个大类算法,这些理论是基础,后面的随机森林,GBDT等等算法都是这些算法的提升。需要着重理解。
5、实现
此处,我们给出ID3算法和CART算法的一些实现,如下所示:
ID3:
from math import log import operator def createDataSet(): dataSet = [[1, 1, 'yes'], [1, 1, 'yes'], [1, 0, 'no'], [0, 1, 'no'], [0, 1, 'no']] labels = ['no surfacing','flippers'] #change to discrete values return dataSet, labels def calcShannonEnt(dataSet): numEntries = len(dataSet) labelCounts = {} for featVec in dataSet: #the the number of unique elements and their occurance currentLabel = featVec[-1] if currentLabel not in labelCounts.keys(): labelCounts[currentLabel] = 0 labelCounts[currentLabel] += 1 shannonEnt = 0.0 for key in labelCounts: prob = float(labelCounts[key])/numEntries shannonEnt -= prob * log(prob,2) #log base 2 return shannonEnt def splitDataSet(dataSet, axis, value): retDataSet = [] for featVec in dataSet: if featVec[axis] == value: reducedFeatVec = featVec[:axis] #chop out axis used for splitting reducedFeatVec.extend(featVec[axis+1:]) retDataSet.append(reducedFeatVec) return retDataSet def chooseBestFeatureToSplit(dataSet): numFeatures = len(dataSet[0]) - 1 #the last column is used for the labels baseEntropy = calcShannonEnt(dataSet) bestInfoGain = 0.0; bestFeature = -1 for i in range(numFeatures): #iterate over all the features featList = [example[i] for example in dataSet]#create a list of all the examples of this feature uniqueVals = set(featList) #get a set of unique values newEntropy = 0.0 for value in uniqueVals: subDataSet = splitDataSet(dataSet, i, value) prob = len(subDataSet)/float(len(dataSet)) newEntropy += prob * calcShannonEnt(subDataSet) infoGain = baseEntropy - newEntropy #calculate the info gain; ie reduction in entropy if (infoGain > bestInfoGain): #compare this to the best gain so far bestInfoGain = infoGain #if better than current best, set to best bestFeature = i return bestFeature #returns an integer def majorityCnt(classList): classCount={} for vote in classList: if vote not in classCount.keys(): classCount[vote] = 0 classCount[vote] += 1 sortedClassCount = sorted(classCount.iteritems(), key=operator.itemgetter(1), reverse=True) return sortedClassCount[0][0] def createTree(dataSet,labels): classList = [example[-1] for example in dataSet] if classList.count(classList[0]) == len(classList): return classList[0]#stop splitting when all of the classes are equal if len(dataSet[0]) == 1: #stop splitting when there are no more features in dataSet return majorityCnt(classList) bestFeat = chooseBestFeatureToSplit(dataSet) bestFeatLabel = labels[bestFeat] myTree = {bestFeatLabel:{}} del(labels[bestFeat]) featValues = [example[bestFeat] for example in dataSet] uniqueVals = set(featValues) for value in uniqueVals: subLabels = labels[:] #copy all of labels, so trees don't mess up existing labels myTree[bestFeatLabel][value] = createTree(splitDataSet(dataSet, bestFeat, value),subLabels) return myTree def classify(inputTree,featLabels,testVec): firstStr = inputTree.keys()[0] secondDict = inputTree[firstStr] featIndex = featLabels.index(firstStr) key = testVec[featIndex] valueOfFeat = secondDict[key] if isinstance(valueOfFeat, dict): classLabel = classify(valueOfFeat, featLabels, testVec) else: classLabel = valueOfFeat return classLabel def storeTree(inputTree,filename): import pickle fw = open(filename,'w') pickle.dump(inputTree,fw) fw.close() def grabTree(filename): import pickle fr = open(filename) return pickle.load(fr)
CART:
''' Created on Feb 4, 2011 Tree-Based Regression Methods @author: Peter Harrington ''' from numpy import * def loadDataSet(fileName): #general function to parse tab -delimited floats dataMat = [] #assume last column is target value fr = open(fileName) for line in fr.readlines(): curLine = line.strip().split('\t') fltLine = map(float,curLine) #map all elements to float() dataMat.append(fltLine) return dataMat def binSplitDataSet(dataSet, feature, value): mat0 = dataSet[nonzero(dataSet[:,feature] > value)[0],:][0] mat1 = dataSet[nonzero(dataSet[:,feature] <= value)[0],:][0] return mat0,mat1 def regLeaf(dataSet):#returns the value used for each leaf return mean(dataSet[:,-1]) def regErr(dataSet): return var(dataSet[:,-1]) * shape(dataSet)[0] def linearSolve(dataSet): #helper function used in two places m,n = shape(dataSet) X = mat(ones((m,n))); Y = mat(ones((m,1)))#create a copy of data with 1 in 0th postion X[:,1:n] = dataSet[:,0:n-1]; Y = dataSet[:,-1]#and strip out Y xTx = X.T*X if linalg.det(xTx) == 0.0: raise NameError('This matrix is singular, cannot do inverse,\n\ try increasing the second value of ops') ws = xTx.I * (X.T * Y) return ws,X,Y def modelLeaf(dataSet):#create linear model and return coeficients ws,X,Y = linearSolve(dataSet) return ws def modelErr(dataSet): ws,X,Y = linearSolve(dataSet) yHat = X * ws return sum(power(Y - yHat,2)) def chooseBestSplit(dataSet, leafType=regLeaf, errType=regErr, ops=(1,4)): tolS = ops[0]; tolN = ops[1] #if all the target variables are the same value: quit and return value if len(set(dataSet[:,-1].T.tolist()[0])) == 1: #exit cond 1 return None, leafType(dataSet) m,n = shape(dataSet) #the choice of the best feature is driven by Reduction in RSS error from mean S = errType(dataSet) bestS = inf; bestIndex = 0; bestValue = 0 for featIndex in range(n-1): for splitVal in set(dataSet[:,featIndex]): mat0, mat1 = binSplitDataSet(dataSet, featIndex, splitVal) if (shape(mat0)[0] < tolN) or (shape(mat1)[0] < tolN): continue newS = errType(mat0) + errType(mat1) if newS < bestS: bestIndex = featIndex bestValue = splitVal bestS = newS #if the decrease (S-bestS) is less than a threshold don't do the split if (S - bestS) < tolS: return None, leafType(dataSet) #exit cond 2 mat0, mat1 = binSplitDataSet(dataSet, bestIndex, bestValue) if (shape(mat0)[0] < tolN) or (shape(mat1)[0] < tolN): #exit cond 3 return None, leafType(dataSet) return bestIndex,bestValue#returns the best feature to split on #and the value used for that split def createTree(dataSet, leafType=regLeaf, errType=regErr, ops=(1,4)):#assume dataSet is NumPy Mat so we can array filtering feat, val = chooseBestSplit(dataSet, leafType, errType, ops)#choose the best split if feat == None: return val #if the splitting hit a stop condition return val retTree = {} retTree['spInd'] = feat retTree['spVal'] = val lSet, rSet = binSplitDataSet(dataSet, feat, val) retTree['left'] = createTree(lSet, leafType, errType, ops) retTree['right'] = createTree(rSet, leafType, errType, ops) return retTree def isTree(obj): return (type(obj).__name__=='dict') def getMean(tree): if isTree(tree['right']): tree['right'] = getMean(tree['right']) if isTree(tree['left']): tree['left'] = getMean(tree['left']) return (tree['left']+tree['right'])/2.0 def prune(tree, testData): if shape(testData)[0] == 0: return getMean(tree) #if we have no test data collapse the tree if (isTree(tree['right']) or isTree(tree['left'])):#if the branches are not trees try to prune them lSet, rSet = binSplitDataSet(testData, tree['spInd'], tree['spVal']) if isTree(tree['left']): tree['left'] = prune(tree['left'], lSet) if isTree(tree['right']): tree['right'] = prune(tree['right'], rSet) #if they are now both leafs, see if we can merge them if not isTree(tree['left']) and not isTree(tree['right']): lSet, rSet = binSplitDataSet(testData, tree['spInd'], tree['spVal']) errorNoMerge = sum(power(lSet[:,-1] - tree['left'],2)) +\ sum(power(rSet[:,-1] - tree['right'],2)) treeMean = (tree['left']+tree['right'])/2.0 errorMerge = sum(power(testData[:,-1] - treeMean,2)) if errorMerge < errorNoMerge: print "merging" return treeMean else: return tree else: return tree def regTreeEval(model, inDat): return float(model) def modelTreeEval(model, inDat): n = shape(inDat)[1] X = mat(ones((1,n+1))) X[:,1:n+1]=inDat return float(X*model) def treeForeCast(tree, inData, modelEval=regTreeEval): if not isTree(tree): return modelEval(tree, inData) if inData[tree['spInd']] > tree['spVal']: if isTree(tree['left']): return treeForeCast(tree['left'], inData, modelEval) else: return modelEval(tree['left'], inData) else: if isTree(tree['right']): return treeForeCast(tree['right'], inData, modelEval) else: return modelEval(tree['right'], inData) def createForeCast(tree, testData, modelEval=regTreeEval): m=len(testData) yHat = mat(zeros((m,1))) for i in range(m): yHat[i,0] = treeForeCast(tree, mat(testData[i]), modelEval) return yHat
