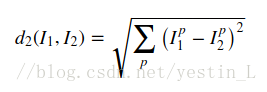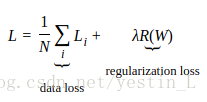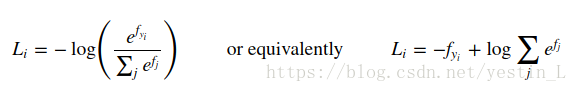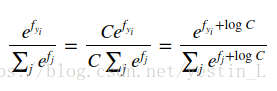一、knn:
1.knn.py中实现
a.分别用双重循环、单重循环和不使用循环实现欧式距离公式
公式:
双重循环:
def compute_distances_two_loops(self, X):
"""
Compute the distance between each test point in X and each training point
in self.X_train using a nested loop over both the training data and the
test data.
Inputs:
- X: A numpy array of shape (num_test, D) containing test data.
Returns:
- dists: A numpy array of shape (num_test, num_train) where dists[i, j]
is the Euclidean distance between the ith test point and the jth training
point.
"""
num_test = X.shape[0]
num_train = self.X_train.shape[0]
dists = np.zeros((num_test, num_train))
for i in xrange(num_test):
for j in xrange(num_train):
#####################################################################
# TODO: #
# Compute the l2 distance between the ith test point and the jth #
# training point, and store the result in dists[i, j]. You should #
# not use a loop over dimension. #
#####################################################################
dists[i,j] = np.sqrt(np.sum(np.square(self.X_train[j,:] - X[i,:])))
#####################################################################
# END OF YOUR CODE #
#####################################################################
return dists单重循环:
def compute_distances_one_loop(self, X):
"""
Compute the distance between each test point in X and each training point
in self.X_train using a single loop over the test data.
Input / Output: Same as compute_distances_two_loops
"""
num_test = X.shape[0]
num_train = self.X_train.shape[0]
dists = np.zeros((num_test, num_train))
for i in xrange(num_test):
#######################################################################
# TODO: #
# Compute the l2 distance between the ith test point and all training #
# points, and store the result in dists[i, :]. #
#######################################################################
dists[i,:] = np.sqrt(np.sum(np.square(self.X_train - X[i,:]),axis = 1))
#######################################################################
# END OF YOUR CODE #
#######################################################################
return dists代码部分
def compute_distances_no_loops(self, X):
"""
Compute the distance between each test point in X and each training point
in self.X_train using no explicit loops.
Input / Output: Same as compute_distances_two_loops
"""
num_test = X.shape[0]
num_train = self.X_train.shape[0]
dists = np.zeros((num_test, num_train))
#########################################################################
# TODO: #
# Compute the l2 distance between all test points and all training #
# points without using any explicit loops, and store the result in #
# dists. #
# #
# You should implement this function using only basic array operations; #
# in particular you should not use functions from scipy. #
# #
# HINT: Try to formulate the l2 distance using matrix multiplication #
# and two broadcast sums. #
#########################################################################
dist1 = np.sum(np.square(self.X_train),axis=1)
dist2 = np.sum(np.square(X),axis=1)
dist3 = np.dot(X,self.X_train.T)
dists = np.sqrt(-2*dist3+dist1+dist2.reshape(dist2.shape[0],1))
#########################################################################
# END OF YOUR CODE #
#########################################################################
return dists公式推导:
此处参考了https://www.2cto.com/kf/201710/693091.html
b.使用knn方法实现类别预测
完整代码:
def predict_labels(self, dists, k=1):
"""
Given a matrix of distances between test points and training points,
predict a label for each test point.
Inputs:
- dists: A numpy array of shape (num_test, num_train) where dists[i, j]
gives the distance betwen the ith test point and the jth training point.
Returns:
- y: A numpy array of shape (num_test,) containing predicted labels for the
test data, where y[i] is the predicted label for the test point X[i].
"""
num_test = dists.shape[0]
y_pred = np.zeros(num_test)
for i in xrange(num_test):
# A list of length k storing the labels of the k nearest neighbors to
# the ith test point.
closest_y = []
#########################################################################
# TODO: #
# Use the distance matrix to find the k nearest neighbors of the ith #
# testing point, and use self.y_train to find the labels of these #
# neighbors. Store these labels in closest_y. #
# Hint: Look up the function numpy.argsort. #
#########################################################################
closest_y = self.y_train[np.argsort(dists[i,:])[:k]]
#########################################################################
# TODO: #
# Now that you have found the labels of the k nearest neighbors, you #
# need to find the most common label in the list closest_y of labels. #
# Store this label in y_pred[i]. Break ties by choosing the smaller #
# label. #
#########################################################################
y_pred[i] = np.argmax(np.bincount(closest_y))
#########################################################################
# END OF YOUR CODE #
#########################################################################
return y_pred 代码解析:
closest_y = self.y_train[np.argsort(dists[i,:])[:k]]用np.argsort函数实现对现在的dists每一行从小到大排序,返回相应的所在切片的值。同时获取训练值中y标签前k个对应切片位置的类别值,并存入用y_pred的相应行中。
y_pred[i] = np.argmax(np.bincount(closest_y))2.knn.ipynb代码实现
Cross Validation
num_folds = 5
k_choices = [1, 3, 5, 8, 10, 12, 15, 20, 50, 100]
X_train_folds = []
y_train_folds = []
################################################################################
# TODO: #
# Split up the training data into folds. After splitting, X_train_folds and #
# y_train_folds should each be lists of length num_folds, where #
# y_train_folds[i] is the label vector for the points in X_train_folds[i]. #
# Hint: Look up the numpy array_split function. #
################################################################################
X_train_folds = np.array_split(X_train,num_folds)
y_train_folds = np.array_split(y_train,num_folds)
################################################################################
# END OF YOUR CODE #
################################################################################
# A dictionary holding the accuracies for different values of k that we find
# when running cross-validation. After running cross-validation,
# k_to_accuracies[k] should be a list of length num_folds giving the different
# accuracy values that we found when using that value of k.
k_to_accuracies = {}
################################################################################
# TODO: #
# Perform k-fold cross validation to find the best value of k. For each #
# possible value of k, run the k-nearest-neighbor algorithm num_folds times, #
# where in each case you use all but one of the folds as training data and the #
# last fold as a validation set. Store the accuracies for all fold and all #
# values of k in the k_to_accuracies dictionary. #
################################################################################
for k in k_choices:
k_to_accuracies[k] = np.zeros(num_folds)
for i in range(num_folds):
Xtr = np.array(X_train_folds[:i]+X_train_folds[i+1:])
Xte = np.array(X_train_folds[i])
ytr = np.array(y_train_folds[:i]+y_train_folds[i+1:])
yte = np.array(y_train_folds[i])
#reshape the training and test array
Xtr = np.reshape(Xtr,(Xtr.shape[0]*Xtr.shape[1],-1))
ytr = np.reshape(ytr,(ytr.shape[0]*ytr.shape[1],-1))
ytr = np.reshape(ytr,len(ytr)) #flatten the shape of ytr into 1*4000
classifier.train(Xtr,ytr)
yte_pred = classifier.predict(Xte,k)
num_correct = np.sum(yte_pred == yte)
accuracy = float(num_correct) / len(yte)
k_to_accuracies[k][i] = accuracy
################################################################################
# END OF YOUR CODE #
################################################################################
# Print out the computed accuracies
for k in sorted(k_to_accuracies):
for accuracy in k_to_accuracies[k]:
print('k = %d, accuracy = %f' % (k, accuracy))参考链接:https://blog.csdn.net/MargretWG/article/details/69056256
代码详解:
X_train_folds = np.array_split(X_train,num_folds)
y_train_folds = np.array_split(y_train,num_folds)用np.array_split函数将训练集和标签均分成5份,用于交叉验证
for k in k_choices: #试k的值,寻找最优k值
k_to_accuracies[k] = np.zeros(num_folds) #让字典中的key值为不同k值,且一个key对应5个值
for i in range(num_folds): #让分类的训练集循环做测试集
Xtr = np.array(X_train_folds[:i]+X_train_folds[i+1:]) #将第i个训练集做测试集,其余做训练集
Xte = np.array(X_train_folds[i]) #Xtr.shape=(4,1000,3072) Xte.shape=(1000,3072)
ytr = np.array(y_train_folds[:i]+y_train_folds[i+1:]) #ytr.shape=(1000,) yte.shape=(4,1000,1)
yte = np.array(y_train_folds[i])
#reshape the training and test array
Xtr = np.reshape(Xtr,(Xtr.shape[0]*Xtr.shape[1],-1)) #Xtr.shape=(4000,3072)
ytr = np.reshape(ytr,(ytr.shape[0]*ytr.shape[1],-1)) #ytr.shape=(4000,1)
ytr = np.reshape(ytr,len(ytr)) #flatten the shape of ytr into 1*4000
#ytr.shape=(4000,)
classifier.train(Xtr,ytr) #输入训练集
yte_pred = classifier.predict(Xte,k) #预测标签并返回,使用无循环
num_correct = np.sum(yte_pred == yte)
accuracy = float(num_correct) / len(yte)
k_to_accuracies[k][i] = accuracy #将准确值存入参考链接中缺了红色字体的部分代码,输出会报错
#ValueError: object too deep for desired array
加上红色部分代码就可以。这是因为如果不加上的话,ytr.shape是一个4000*1的二维矩阵,我试过输入代码
ytr = np.reshape(ytr,(-1,ytr.shape[0]*ytr.shape[1]))这样也不行,虽然这样返回的是一个1*4000的矩阵,但这个在numpy里会被认为是二维的矩阵,而不是我们直观上的一维行向量。如果用print(ytr) ,会等于[ [ ....] ] 的一个二维矩阵。只有再reshape(ytr,len(ytr))时才能将二维矩阵化为一个一维的,这时再print(ytr),就会输出[...]的一个一维行向量。
而且还要注意的一点是,len()返回的是矩阵的行数量,所以这里要先把ytr变成4000*1的二维矩阵,而非1*4000的。
总之一定要注意输入参数的维度。
二、SVM
1.linear_svm.py代码实现及推导
a.svm_loss_naive
loss部分比较简单,代码已经给出,就是完成公式
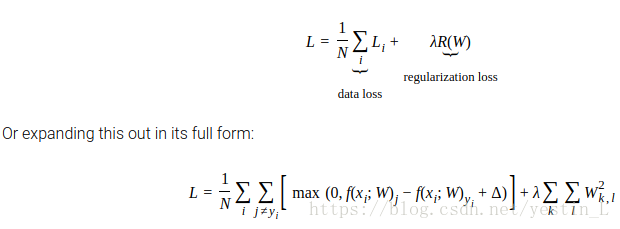
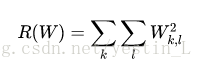
计算gradient并存入dW中
代码:
def svm_loss_naive(W, X, y, reg):
"""
Structured SVM loss function, naive implementation (with loops).
Inputs have dimension D, there are C classes, and we operate on minibatches
of N examples.
Inputs:
- W: A numpy array of shape (D, C) containing weights.
- X: A numpy array of shape (N, D) containing a minibatch of data.
- y: A numpy array of shape (N,) containing training labels; y[i] = c means
that X[i] has label c, where 0 <= c < C.
- reg: (float) regularization strength
Returns a tuple of:
- loss as single float
- gradient with respect to weights W; an array of same shape as W
"""
dW = np.zeros(W.shape) # initialize the gradient as zero
# compute the loss and the gradient
num_classes = W.shape[1]
num_train = X.shape[0]
loss = 0.0
for i in xrange(num_train):
scores = X[i].dot(W)
correct_class_score = scores[y[i]]
for j in xrange(num_classes):
if j == y[i]:
continue
margin = scores[j] - correct_class_score + 1 # note delta = 1
if margin > 0:
loss += margin
dW[:,y[i]] += -X[i,:].T
dW[:,j] += X[i,].T
# Right now the loss is a sum over all training examples, but we want it
# to be an average instead so we divide by num_train.
loss /= num_train
dW /= num_train
# Add regularization to the loss.
loss += reg * np.sum(W * W)
dW += reg*W推导
b.svm_loss_vectorized
def svm_loss_vectorized(W, X, y, reg):
"""
Structured SVM loss function, vectorized implementation.
Inputs and outputs are the same as svm_loss_naive.
"""
loss = 0.0
dW = np.zeros(W.shape) # initialize the gradient as zero
num_train = X.shape[0]
#############################################################################
# TODO: #
# Implement a vectorized version of the structured SVM loss, storing the #
# result in loss. #
#############################################################################
scores = X.dot(W)
correct_class_score = scores[np.arange(num_train), y]
correct_class_score = np.reshape(correct_class_score,(num_train,-1))
margins = scores - correct_class_score + 1.0
margins[np.arange(num_train),y] = 0.0
margins[margins <= 0] = 0.0
loss += np.sum(margins) / num_train
loss += reg*np.sum(W*W)
#############################################################################
# END OF YOUR CODE #
#############################################################################
#############################################################################
# TODO: #
# Implement a vectorized version of the gradient for the structured SVM #
# loss, storing the result in dW. #
# #
# Hint: Instead of computing the gradient from scratch, it may be easier #
# to reuse some of the intermediate values that you used to compute the #
# loss. #
#############################################################################
margins[margins > 0] = 1.0
row_sum = np.sum(margins,axis=1)
margins[np.arange(num_train), y] = -row_sum
dW += np.dot(X.T, margins) / num_train + reg*W
#############################################################################
# END OF YOUR CODE #
#############################################################################
return loss, dW原理同上,这里只是去掉了循环,将操作矩阵化了,使代码运行效率更高
2.linear_classifier.py代码实现及推导
a.train函数
训练后得到权重矩阵存入self.W中
代码
def train(self, X, y, learning_rate=1e-3, reg=1e-5, num_iters=100,
batch_size=200, verbose=False):
"""
Train this linear classifier using stochastic gradient descent.
Inputs:
- X: A numpy array of shape (N, D) containing training data; there are N
training samples each of dimension D.
- y: A numpy array of shape (N,) containing training labels; y[i] = c
means that X[i] has label 0 <= c < C for C classes.
- learning_rate: (float) learning rate for optimization.
- reg: (float) regularization strength.
- num_iters: (integer) number of steps to take when optimizing
- batch_size: (integer) number of training examples to use at each step.
- verbose: (boolean) If true, print progress during optimization.
Outputs:
A list containing the value of the loss function at each training iteration.
"""
num_train, dim = X.shape
num_classes = np.max(y) + 1 # assume y takes values 0...K-1 where K is number of classes
if self.W is None:
# lazily initialize W
self.W = 0.001 * np.random.randn(dim, num_classes)
# Run stochastic gradient descent to optimize W
loss_history = []
for it in xrange(num_iters):
X_batch = None
y_batch = None
#########################################################################
# TODO: #
# Sample batch_size elements from the training data and their #
# corresponding labels to use in this round of gradient descent. #
# Store the data in X_batch and their corresponding labels in #
# y_batch; after sampling X_batch should have shape (dim, batch_size) #
# and y_batch should have shape (batch_size,) #
# #
# Hint: Use np.random.choice to generate indices. Sampling with #
# replacement is faster than sampling without replacement. #
#########################################################################
sample_index = np.random.choice(num_train,batch_size,replace = False)
X_batch = X[sample_index,:]
y_batch = y[sample_index]
#########################################################################
# END OF YOUR CODE #
#########################################################################
# evaluate loss and gradient
loss, grad = self.loss(X_batch, y_batch, reg)
loss_history.append(loss)
# perform parameter update
#########################################################################
# TODO: #
# Update the weights using the gradient and the learning rate. #
#########################################################################
self.W -= learning_rate*grad
#########################################################################
# END OF YOUR CODE #
#########################################################################
if verbose and it % 100 == 0:
print('iteration %d / %d: loss %f' % (it, num_iters, loss))
return loss_history代码解析:
sample_index = np.random.choice(num_train,batch_size,replace = False)
X_batch = X[sample_index,:]
y_batch = y[sample_index]用np.random.choice函数从500个训练集中随机挑选batch_size个样本及其标签
self.W -= learning_rate*grad更新权重值
b.predict
代码:
def predict(self, X):
"""
Use the trained weights of this linear classifier to predict labels for
data points.
Inputs:
- X: A numpy array of shape (N, D) containing training data; there are N
training samples each of dimension D.
Returns:
- y_pred: Predicted labels for the data in X. y_pred is a 1-dimensional
array of length N, and each element is an integer giving the predicted
class.
"""
y_pred = np.zeros(X.shape[0])
###########################################################################
# TODO: #
# Implement this method. Store the predicted labels in y_pred. #
###########################################################################
score = X.dot(self.W)
y_pred = np.argmax(score,axis=1)
###########################################################################
# END OF YOUR CODE #
###########################################################################
return y_pred将X与W相乘获得相应的score,然后获取最大分数所对应的类别
3.svm.ipynb
用validation集去测试训练集所获得的hyperparameters,并记录相应得分,取最高得分的hypermeters作为最终的参数
# Use the validation set to tune hyperparameters (regularization strength and
# learning rate). You should experiment with different ranges for the learning
# rates and regularization strengths; if you are careful you should be able to
# get a classification accuracy of about 0.4 on the validation set.
learning_rates = [1.8e-7, 1.9e-7, 2e-7, 2.1e-7, 2.2e-7]
regularization_strengths = [2.3e4,2.4e4,2.5e4, 2.6e4, 2.7e4]
# results is dictionary mapping tuples of the form
# (learning_rate, regularization_strength) to tuples of the form
# (training_accuracy, validation_accuracy). The accuracy is simply the fraction
# of data points that are correctly classified.
results = {}
best_val = -1 # The highest validation accuracy that we have seen so far.
best_svm = None # The LinearSVM object that achieved the highest validation rate.
################################################################################
# TODO: #
# Write code that chooses the best hyperparameters by tuning on the validation #
# set. For each combination of hyperparameters, train a linear SVM on the #
# training set, compute its accuracy on the training and validation sets, and #
# store these numbers in the results dictionary. In addition, store the best #
# validation accuracy in best_val and the LinearSVM object that achieves this #
# accuracy in best_svm. #
# #
# Hint: You should use a small value for num_iters as you develop your #
# validation code so that the SVMs don't take much time to train; once you are #
# confident that your validation code works, you should rerun the validation #
# code with a larger value for num_iters. #
################################################################################
iters = 3000
for lr in learning_rates:
for rs in regularization_strengths:
svm.train(X_train, y_train, learning_rate=lr, reg=rs, num_iters=iters)
y_train_pred = svm.predict(X_train)
accu_train = np.mean(y_train==y_train_pred)
y_val_pred = svm.predict(X_val)
accu_val = np.mean(y_val==y_val_pred)
results[(lr,rs)] = (accu_train, accu_val)
if best_val < accu_val:
best_val = accu_val
best_svm = svm
################################################################################
# END OF YOUR CODE #
################################################################################
# Print out results.
for lr, reg in sorted(results):
train_accuracy, val_accuracy = results[(lr, reg)]
print('lr %e reg %e train accuracy: %f val accuracy: %f' % (
lr, reg, train_accuracy, val_accuracy))
print('best validation accuracy achieved during cross-validation: %f' % best_val)最后得到的最优准确度在3.9~4.0左右。
三、softmax
1.softmax.py
a.softmax_loss_naive函数代码实现及推导
代码:
def softmax_loss_naive(W, X, y, reg):
"""
Softmax loss function, naive implementation (with loops)
Inputs have dimension D, there are C classes, and we operate on minibatches
of N examples.
Inputs:
- W: A numpy array of shape (D, C) containing weights.
- X: A numpy array of shape (N, D) containing a minibatch of data.
- y: A numpy array of shape (N,) containing training labels; y[i] = c means
that X[i] has label c, where 0 <= c < C.
- reg: (float) regularization strength
Returns a tuple of:
- loss as single float
- gradient with respect to weights W; an array of same shape as W
"""
# Initialize the loss and gradient to zero.
loss = 0.0
dW = np.zeros_like(W)
#############################################################################
# TODO: Compute the softmax loss and its gradient using explicit loops. #
# Store the loss in loss and the gradient in dW. If you are not careful #
# here, it is easy to run into numeric instability. Don't forget the #
# regularization! #
#############################################################################
num_train = X.shape[0]
num_classes = W.shape[1]
for i in xrange(num_train):
scores = X[i].dot(W)
scores -= np.max(scores)
correct_class_score = scores[y[i]]
loss += -correct_class_score + np.log(np.sum(np.exp(scores)))
dW[:,y[i]] -= X[i].T
for j in xrange(num_classes):
dW[:,j] += X[i].T.dot(np.exp(scores[j])/(np.sum(np.exp(scores))))
loss /= num_train
loss += reg*np.sum(W*W)
dW /= num_train
dW += reg*W
#############################################################################
# END OF YOUR CODE #
#############################################################################
return loss, dW代码解析:
loss函数实现公式
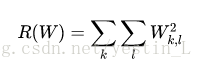
loss函数的代码比较好理解,只是要注意归一化,这里用了所有值减去最大值的办法
令
难点在于dW的求解
此处参考了https://www.jianshu.com/p/6e405cecd609 ,我修改了一下原作中一些不太清晰的下标
b.softmax_loss_vectorized代码实现和推导
代码:
def softmax_loss_vectorized(W, X, y, reg):
"""
Softmax loss function, vectorized version.
Inputs and outputs are the same as softmax_loss_naive.
"""
# Initialize the loss and gradient to zero.
loss = 0.0
dW = np.zeros_like(W)
#############################################################################
# TODO: Compute the softmax loss and its gradient using no explicit loops. #
# Store the loss in loss and the gradient in dW. If you are not careful #
# here, it is easy to run into numeric instability. Don't forget the #
# regularization! #
#############################################################################
num_train = X.shape[0]
num_classes = W.shape[1]
scores = X.dot(W)
scores_row_max = np.max(scores, axis=1).reshape(-1,1)
scores -= scores_row_max
correct_class_scores = scores[np.arange(num_train), y]
loss += -np.sum(correct_class_scores) + np.sum(np.log(np.sum(np.exp(scores),axis=1)))
loss /= num_train
loss += reg*np.sum(W*W)
Pj = np.exp(scores)/(np.sum(np.exp(scores), axis=1).reshape(-1,1))
Pj[range(num_train), y] -= 1
dW = X.T.dot(Pj)
dW /= num_train
dW += reg*W
#############################################################################
# END OF YOUR CODE #
#############################################################################
return loss, dW代码解析:
loss函数的实现相对简单,dW的矩阵化计算较为巧妙,实现公式
if j==yi 则 dW = X*(Pj -1)
j!=yi 则 dW = X*Pj
其中Pj为
2.softmax.ipynb
用validation调整参数部分代码
# Use the validation set to tune hyperparameters (regularization strength and
# learning rate). You should experiment with different ranges for the learning
# rates and regularization strengths; if you are careful you should be able to
# get a classification accuracy of over 0.35 on the validation set.
from cs231n.classifiers import Softmax
results = {}
best_val = -1
best_softmax = None
learning_rates = [5e-7,5.5e7, 6e-7]
regularization_strengths = [5e4, 5.5e4]
################################################################################
# TODO: #
# Use the validation set to set the learning rate and regularization strength. #
# This should be identical to the validation that you did for the SVM; save #
# the best trained softmax classifer in best_softmax. #
################################################################################
Softmax = Softmax()
iters = 500
for lr in learning_rates:
for rs in regularization_strengths:
Softmax.train(X_train, y_train, learning_rate=lr, reg=rs, num_iters=iters)
y_train_pred = Softmax.predict(X_train)
accu_train = np.mean(y_train==y_train_pred)
y_val_pred = Softmax.predict(X_val)
accu_val = np.mean(y_val==y_val_pred)
results[(lr,rs)] = (accu_train, accu_val)
if best_val < accu_val:
best_softmax = Softmax
best_val = accu_val
################################################################################
# END OF YOUR CODE #
################################################################################
# Print out results.
for lr, reg in sorted(results):
train_accuracy, val_accuracy = results[(lr, reg)]
print('lr %e reg %e train accuracy: %f val accuracy: %f' % (
lr, reg, train_accuracy, val_accuracy))
print('best validation accuracy achieved during cross-validation: %f' % best_val)此处和svm的一样,记得第一行要进行Sotfmax类的实例化
四、Two_layer_net
1.neutral_net.py
a.loss函数实现和推导
代码:
def loss(self, X, y=None, reg=0.0):
"""
Compute the loss and gradients for a two layer fully connected neural
network.
Inputs:
- X: Input data of shape (N, D). Each X[i] is a training sample.
- y: Vector of training labels. y[i] is the label for X[i], and each y[i] is
an integer in the range 0 <= y[i] < C. This parameter is optional; if it
is not passed then we only return scores, and if it is passed then we
instead return the loss and gradients.
- reg: Regularization strength.
Returns:
If y is None, return a matrix scores of shape (N, C) where scores[i, c] is
the score for class c on input X[i].
If y is not None, instead return a tuple of:
- loss: Loss (data loss and regularization loss) for this batch of training
samples.
- grads: Dictionary mapping parameter names to gradients of those parameters
with respect to the loss function; has the same keys as self.params.
"""
# Unpack variables from the params dictionary
W1, b1 = self.params['W1'], self.params['b1']
W2, b2 = self.params['W2'], self.params['b2']
N, D = X.shape
# Compute the forward pass
scores = None
#############################################################################
# TODO: Perform the forward pass, computing the class scores for the input. #
# Store the result in the scores variable, which should be an array of #
# shape (N, C). #
#############################################################################
f = lambda x: np.maximum(0,x)
# f = lambda x: np.tanh(x)
# f = lambda x: 1/(1+np.exp(x))
a1 = np.dot(X,W1) + b1
h1 = f(a1)
a2 = np.dot(h1,W2) + b2
scores = a2
#############################################################################
# END OF YOUR CODE #
#############################################################################
# If the targets are not given then jump out, we're done
if y is None:
return scores
# Compute the loss
loss = None
#############################################################################
# TODO: Finish the forward pass, and compute the loss. This should include #
# both the data loss and L2 regularization for W1 and W2. Store the result #
# in the variable loss, which should be a scalar. Use the Softmax #
# classifier loss. #
#############################################################################
scores_row_max = np.max(scores, axis=1).reshape(-1,1)
scores -= scores_row_max
correct_class_scores = scores[np.arange(N), y]
loss = -np.sum(correct_class_scores) + np.sum(np.log(np.sum(np.exp(scores), axis=1)))
loss /= N
loss += reg * (np.sum(W1 * W1)+np.sum(W2*W2))
#############################################################################
# END OF YOUR CODE #
#############################################################################
# Backward pass: compute gradients
grads = {}
#############################################################################
# TODO: Compute the backward pass, computing the derivatives of the weights #
# and biases. Store the results in the grads dictionary. For example, #
# grads['W1'] should store the gradient on W1, and be a matrix of same size #
#############################################################################
P = np.exp(scores) / (np.sum(np.exp(scores), axis=1).reshape(-1, 1))
dscores = P
dscores[range(N),y] -= 1
dscores /= N
dW2 = np.dot(h1.T, dscores) + 2*reg*W2
db2 = np.sum(dscores, axis=0, keepdims=False)
da2 = np.dot(dscores,W2.T)
da2[a1 < 0] = 0
dW1 = np.dot(X.T,da2) + 2*reg*W1
db1 = np.sum(da2, axis=0, keepdims=False)
grads['W1'] = dW1
grads['b1'] = db1
grads['W2'] = dW2
grads['b2'] = db2
#############################################################################
# END OF YOUR CODE #
#############################################################################
return loss, gradspar1. forward pass
# Compute the forward pass
scores = None
#############################################################################
# TODO: Perform the forward pass, computing the class scores for the input. #
# Store the result in the scores variable, which should be an array of #
# shape (N, C). #
#############################################################################
f = lambda x: np.maximum(0,x)
# f = lambda x: np.tanh(x)
# f = lambda x: 1/(1+np.exp(x))
a1 = np.dot(X,W1) + b1
h1 = f(a1)
a2 = np.dot(h1,W2) + b2
scores = a2
#############################################################################
# END OF YOUR CODE #
#############################################################################
# If the targets are not given then jump out, we're done
if y is None:
return scores代码解释:
用Relu激活函数,注释掉的两行是用了logsitic函数和tanh函数试了,效果远不如Relu函数的好,剩下的就是前向传播计算
part2. loss
# Compute the loss
loss = None
#############################################################################
# TODO: Finish the forward pass, and compute the loss. This should include #
# both the data loss and L2 regularization for W1 and W2. Store the result #
# in the variable loss, which should be a scalar. Use the Softmax #
# classifier loss. #
#############################################################################
scores_row_max = np.max(scores, axis=1).reshape(-1,1)
scores -= scores_row_max
correct_class_scores = scores[np.arange(N), y]
loss = -np.sum(correct_class_scores) + np.sum(np.log(np.sum(np.exp(scores), axis=1)))
loss /= N
loss += reg * (np.sum(W1 * W1)+np.sum(W2*W2))
#############################################################################
# END OF YOUR CODE #
#############################################################################代码解释:
先将scores归一化,然后计算使用了softmax分类器后的loss function
part3. Backward pass: compute gradients
# Backward pass: compute gradients
grads = {}
#############################################################################
# TODO: Compute the backward pass, computing the derivatives of the weights #
# and biases. Store the results in the grads dictionary. For example, #
# grads['W1'] should store the gradient on W1, and be a matrix of same size #
#############################################################################
P = np.exp(scores) / (np.sum(np.exp(scores), axis=1).reshape(-1, 1))
dscores = P
dscores[range(N),y] -= 1
dscores /= N
dW2 = np.dot(h1.T, dscores) + 2*reg*W2
db2 = np.sum(dscores, axis=0, keepdims=False)
da2 = np.dot(dscores,W2.T)
da2[a1 < 0] = 0
dW1 = np.dot(X.T,da2) + 2*reg*W1
db1 = np.sum(da2, axis=0, keepdims=False)
grads['W1'] = dW1
grads['b1'] = db1
grads['W2'] = dW2
grads['b2'] = db2
#############################################################################
# END OF YOUR CODE #
#############################################################################
return loss, grads梯度推导:
part4. train(use SGD)
def train(self, X, y, X_val, y_val,
learning_rate=1e-3, learning_rate_decay=0.95,
reg=5e-6, num_iters=100,
batch_size=200, verbose=False):
"""
Train this neural network using stochastic gradient descent.
Inputs:
- X: A numpy array of shape (N, D) giving training data.
- y: A numpy array f shape (N,) giving training labels; y[i] = c means that
X[i] has label c, where 0 <= c < C.
- X_val: A numpy array of shape (N_val, D) giving validation data.
- y_val: A numpy array of shape (N_val,) giving validation labels.
- learning_rate: Scalar giving learning rate for optimization.
- learning_rate_decay: Scalar giving factor used to decay the learning rate
after each epoch.
- reg: Scalar giving regularization strength.
- num_iters: Number of steps to take when optimizing.
- batch_size: Number of training examples to use per step.
- verbose: boolean; if true print progress during optimization.
"""
num_train = X.shape[0]
iterations_per_epoch = max(num_train / batch_size, 1)
# Use SGD to optimize the parameters in self.model
loss_history = []
train_acc_history = []
val_acc_history = []
for it in xrange(num_iters):
X_batch = None
y_batch = None
#########################################################################
# TODO: Create a random minibatch of training data and labels, storing #
# them in X_batch and y_batch respectively. #
#########################################################################
sample_index = np.random.choice(num_train,batch_size)
X_batch = X[sample_index]
y_batch = y[sample_index]
#########################################################################
# END OF YOUR CODE #
#########################################################################
# Compute loss and gradients using the current minibatch
loss, grads = self.loss(X_batch, y=y_batch, reg=reg)
loss_history.append(loss)
#########################################################################
# TODO: Use the gradients in the grads dictionary to update the #
# parameters of the network (stored in the dictionary self.params) #
# using stochastic gradient descent. You'll need to use the gradients #
# stored in the grads dictionary defined above. #
#########################################################################
self.params['W1'] -= learning_rate*grads['W1']
self.params['b1'] -= learning_rate*grads['b1']
self.params['W2'] -= learning_rate*grads['W2']
self.params['b2'] -= learning_rate*grads['b2']
#########################################################################
# END OF YOUR CODE #
#########################################################################
if verbose and it % 100 == 0:
print('iteration %d / %d: loss %f' % (it, num_iters, loss))
# Every epoch, check train and val accuracy and decay learning rate.
if it % iterations_per_epoch == 0:
# Check accuracy
train_acc = (self.predict(X_batch) == y_batch).mean()
val_acc = (self.predict(X_val) == y_val).mean()
train_acc_history.append(train_acc)
val_acc_history.append(val_acc)
# Decay learning rate
learning_rate *= learning_rate_decay
return {
'loss_history': loss_history,
'train_acc_history': train_acc_history,
'val_acc_history': val_acc_history,
}类似前面的linear_classifier中的代码,相对简单
part6.predict
def predict(self, X):
"""
Use the trained weights of this two-layer network to predict labels for
data points. For each data point we predict scores for each of the C
classes, and assign each data point to the class with the highest score.
Inputs:
- X: A numpy array of shape (N, D) giving N D-dimensional data points to
classify.
Returns:
- y_pred: A numpy array of shape (N,) giving predicted labels for each of
the elements of X. For all i, y_pred[i] = c means that X[i] is predicted
to have class c, where 0 <= c < C.
"""
y_pred = None
###########################################################################
# TODO: Implement this function; it should be VERY simple! #
###########################################################################
f = lambda x: np.maximum(0, x)
a1 = np.dot(X, self.params['W1']) + self.params['b1']
h1 = f(a1)
a2 = np.dot(h1, self.params['W2']) + self.params['b2']
scores = a2
y_pred = np.argmax(scores,axis=1)
###########################################################################
# END OF YOUR CODE #
###########################################################################
return y_pred用更新的值去计算最后的类别