Introduction
In this code challenge, we will implement the PCA, an unsupervised dimensionality reduction method. PCA has a very wide range of applications in terms of data analysis and data visualization. Therefore we believe it would bring extra benefit if you can implement PCA from scratch by yourself. Before the code challenge, there is one important note: we only request you to provide minimal implementation during the code challenge. It doesn't necessarily apply that this is how PCA is implemented in the industry. If you are interested, please visit the sklearn official open-source code.
Principle Component Analysis
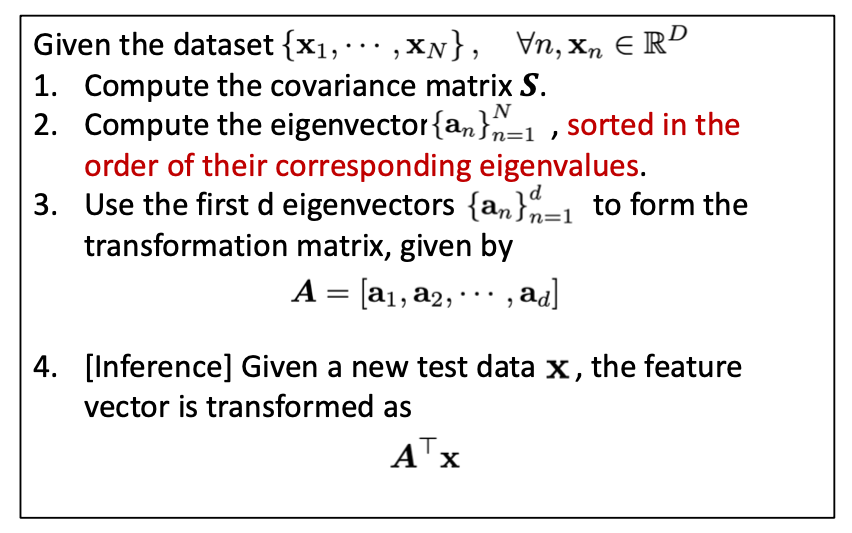
In PCA, we want to find a linear mapping to project the original high-dimensional data to a lower-dimension space so that the maximal information of the data is preserved. Remember as we have covered in the lecture, the target of preserving maximal information, when translated into the language of mathematics, is represented by the following two rules:
-
Dimension is independent from each other.
-
The projected data has the maximal variance.
By solving the optimization problem, we know that the transform matrix of PCA is composed of the eigenvectors of the covariance matrix whose corresponding eigenvalues are sorted from the largest to the smallest.
Here is the pseudo-code of PCA as we already discussed in the lecture:
Instruction
Here is a list of the functions you need to implement:
-
PCA.fit(X): fit the data X, learn the transform matrix self.eig_vecs.
-
PCA.transform(X, n_components=1): transform the data X using the learned transform matrix self.eig_vecs.
-
PCA.fit_transform(X, n_components=1): fit the data X, and return the transformed result of X.
Here is a short example of how our implemented PCA class is used in the real-world application:
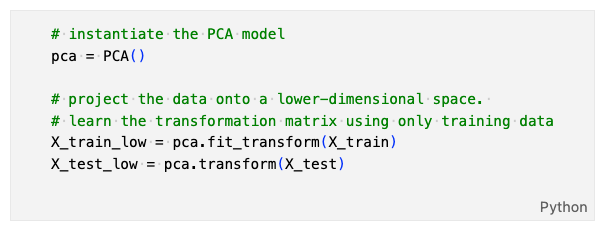
where we first create a LinearReg object, then fit it to the training data, and calculate its loss value for the training data and the test data.
import numpy as np
class PCA(object):
def __init__(self):
# Initialize the eigenvectors and eigenvalues
self.eig_vals, self.eig_vecs = None, None
def fit(self, X):
# implement the .fit() following the algorithm described during the lecture.
# HINT (PCA pseudo code):
# 1. zero-mean the input X;
# 2. compute the covariance matrix
# 3. compute the eigen values and eigen vectors using function **np.linalg.eig()**
# 4. sort the eigen values and eigenvectors using function **np.argsort()**.
# 5. store the eigen vectors
# zero-mean the input X
X_centered = X - np.mean(X, axis=0)
# compute the covariance matrix
cov_matrix = np.cov(X_centered.T)
# compute the eigen values and eigen vectors using np.linalg.eig()
eig_vals, eig_vecs = np.linalg.eig(cov_matrix)
# sort the eigen values and eigenvectors in descending order
sorted_idx = np.argsort(eig_vals)[::-1]
self.eig_vals = eig_vals[sorted_idx]
self.eig_vecs = eig_vecs[:, sorted_idx]
pass
def transform(self, X, n_components=1):
# implement the .transform() using the parameters learned by .fit()
# NOTE: don't forget to 0-mean the input X first.
# Check that we have learned eigenvectors and eigenvalues
assert self.eig_vecs is not None and self.eig_vals is not None, "Fit the model first!"
# 0-mean the input X
X_centered = X - np.mean(X, axis=0)
# Project the centered data onto the principal components
P = self.eig_vecs[:, :n_components]
X_pca = np.dot(X_centered, P)
return X_pca
pass
def fit_transform(self, X, n_components=1):
# implement the PCA transformation based on the .fit() and .transform()
self.fit(X)
return self.transform(X, n_components)
pass