一:k-means、混合高斯聚类的原理
k-means算法的基本思想为:在数据集中根据一定策略选择k个点作为每个簇的初始中心,然后观察剩余的数据,将数据划分到距离这k个点最近的簇中,也就是说将数据划分成k个簇完成一次划分,但形成的新簇并不一定是最好的划分,因此生成的新簇中,重新计算每个簇的中心点,然后在重新进行划分,直到每次划分的结果保持不变。
高斯混合聚类是一种基于概率分布的算法,它首先假设每个簇符合不同的高斯分布,也就是多元正态分布。大致流程为首先假设k个高斯分布,然后判断每个样本属于每个高斯分布的概率,将该样本划分为概率最大的那个分布簇内,然后一轮后更新高斯分布的参数,然后再基于新的参数计算新的分布概率,不断迭代直至模型收敛至局部最优解。
高斯概率函数为:
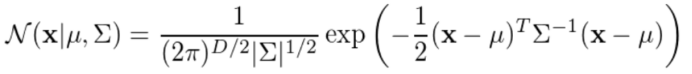
随后计算后验概率:
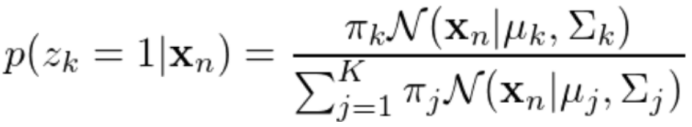
记后验概率为Y(znk),则参数更新规则为:
混合系数: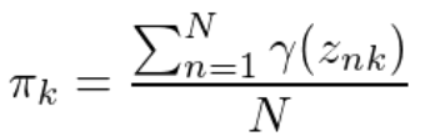
均值向量: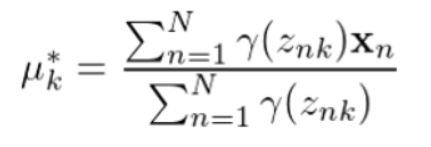
协方差矩阵: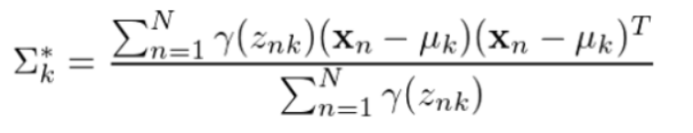
二:程序清单
(一)K-Means聚类:
import random
import pandas as pd
import numpy as np
from itertools import chain
import matplotlib.pyplot as plt
from sklearn import metrics
from sklearn.preprocessing import LabelEncoder
# 读取数据
def getData():
iris = pd.read_csv('./iris.csv',header = None,names=["Sepal.Length","Sepal.Width","Petal.Length","Petal.Width","species"])
X = iris['Sepal.Length'].copy()
X = np.array(X).reshape(-1,1)
Y = iris['Petal.Length'].copy()
Y = np.array(Y).reshape(-1,1)
label = iris['species'].copy()
enc = LabelEncoder() #获取一个LabelEncoder
enc = enc.fit(['Iris-setosa','Iris-versicolor','Iris-virginica']) #训练LabelEncoder
label = enc.transform(label) #使用训练好的LabelEncoder对原数据进行编码
label = np.array(label).reshape(-1,1)
dataSet = np.hstack((X, Y,label))
return dataSet
# 计算欧拉距离
def calcDis(dataSet, centroids, k):
clalist = []
for data in dataSet:
diff = np.tile(data, (k, 1)) - centroids # 分别与三个质心相减
squaredDiff = diff ** 2
squaredDist = np.sum(squaredDiff, axis = 1) # (x1-x2)^2 + (y1-y2)^2
distance = squaredDist ** 0.5
clalist.append(distance)
clalist = np.array(clalist)
return clalist
# 计算质心
def classify(dataSet, centroids, k):
# 计算样本到质心的距离
clalist = calcDis(dataSet, centroids, k)
# 分组并计算新的质
minDistIndices = np.argmin(clalist, axis=1)
newCentroids = (pd.DataFrame(dataSet).groupby(minDistIndices)).mean()
newCentroids = newCentroids.values
# 计算变化量
change = newCentroids - centroids
return change, newCentroids
# 使用K-means分类
def kmeans(dataSet, k):
#随机选取质心
centroids = random.sample(list(dataSet[:,:2]), k)
# 更新质心,直到变化量全为0
change, newCentroids = classify(dataSet[:,:2], centroids, k)
while np.any(change != 0):
change, newCentroids = classify(dataSet[:,:2], newCentroids, k)
centroids = sorted(newCentroids.tolist())
# 根据质心计算每个集群
cluster = []
curr = 0
clalist = calcDis(dataSet[:,:2],centroids,k) # 计算每个点分别到三个质心的距离
minDistIndices = np.argmin(clalist,axis = 1) # 选取距离每个质心距离最小的点返回下标
for i in range(len(minDistIndices)):
if minDistIndices[i] == dataSet[i:i+1,2]:
curr += 1
for i in range(k):
cluster.append([])
for i, j in enumerate(minDistIndices): # 分类
cluster[j].append(dataSet[i:i+1,:2])
return centroids,cluster,minDistIndices,curr
# 可视化结果
def Draw(centroids,cluster):
cluster0 = list(chain.from_iterable(cluster[0]))# 类别1
cluster0 = np.array(cluster0)
cluster1 = list(chain.from_iterable(cluster[1]))# 类别2
cluster1 = np.array(cluster1)
cluster2 = list(chain.from_iterable(cluster[2]))# 类别3
cluster2 = np.array(cluster2)
ax = plt.subplot()
ax.scatter(cluster0[:,0],cluster0[:,1],marker = '*',c = 'green',label = 'Iris-setosa')
ax.scatter(cluster1[:,0],cluster1[:,1],marker = 'o',c = 'blue',label = 'Iris-versicolor')
ax.scatter(cluster2[:,0],cluster2[:,1],marker = '+',c = 'orange',label = 'Iris-virginica')
ax.scatter(centroids[:,0],centroids[:,1],marker='x',c = 'red',label = 'centroid')
plt.title("k-means clustering results")
plt.xlabel("Sepal.Length")
plt.ylabel("Petal.Length")
plt.legend()
plt.show()
def eva_index(dataSet):
# 存放不同的SSE值
sse_list = []
# 轮廓系数
silhouettes = []
for k in range(2,9): # k取值范围
sum = 0
centroids, cluster, minDistance, curr = kmeans(dataset, k)
minDistance = np.array(minDistance)
centroids = np.array(centroids)
# 计算误方差值
for i in range(len(cluster)):
temp = np.sum((cluster[i] - centroids[i])**2)
sum += temp
sse_list.append(sum)
# 计算轮廓系数
silhouette = metrics.silhouette_score(dataSet[:,:2],minDistance,metric='euclidean')
silhouettes.append(silhouette)
# 绘制簇内误方差曲线
plt.subplot(211)
plt.title('KMeans Intra-cluster error variance')
plt.plot(range(2, 9), sse_list, marker='*')
plt.xlabel('Number of clusters')
plt.ylabel('Intra-cluster error variance(SSE)')
plt.show()
# 绘制轮廓系数曲线
plt.subplot(212)
plt.title('KMeans Profile coefficient curve')
plt.plot(range(2,9), silhouettes, marker = '+')
plt.xlabel('Number of clusters')
plt.ylabel('silhouette coefficient')
plt.show()
if __name__=='__main__':
dataset = getData() # 加载数据
eva_index(dataset) # kmeans评价指标
centroids, cluster,minDistance, curr = kmeans(dataset, 3) # k=3时进行聚类
centroids = np.array(centroids)
currD = curr/len(dataset) * 100 # 计算正确率
print("k-means正确率:",currD,"%")
Draw(centroids,cluster) # 结果可视化
二:高斯混合聚类(GMM)
import random
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.preprocessing import LabelEncoder
# 预处理数据
def loadDataSet():
iris = pd.read_csv('./iris.csv',header = None,names=["Sepal.Length","Sepal.Width","Petal.Length","Petal.Width","species"])
X = iris['Sepal.Length'].copy()
X = np.array(X).reshape(-1,1)
Y = iris['Petal.Length'].copy()
Y = np.array(Y).reshape(-1,1)
dataSet = np.hstack((X, Y))
label = iris['species'].copy()
enc = LabelEncoder() #获取一个LabelEncoder
enc = enc.fit(['Iris-setosa','Iris-versicolor','Iris-virginica']) #训练LabelEncoder
label = enc.transform(label) #使用训练好的LabelEncoder对原数据进行编码
return dataSet,label
# 计算欧拉距离
def calcDis(dataSet, centroids, k):
clalist = []
for data in dataSet:
diff = np.tile(data, (k, 1)) - centroids # 分别与三个质心相减
squaredDiff = diff ** 2
squaredDist = np.sum(squaredDiff, axis = 1) # (x1-x2)^2 + (y1-y2)^2
distance = squaredDist ** 0.5
clalist.append(distance)
clalist = np.array(clalist)
return clalist
# 计算质心
def classify(dataSet, centroids, k):
# 计算样本到质心的距离
clalist = calcDis(dataSet, centroids, k)
# 分组并计算新的质心
minDistIndices = np.argmin(clalist, axis=1)
newCentroids = pd.DataFrame(dataSet).groupby(minDistIndices).mean()
newCentroids = newCentroids.values
# 计算变化量
change = newCentroids - centroids
return change, newCentroids
# 使用K-means分类
def kmeans(dataSet, k):
#随机选取质心
centroids = random.sample(list(dataSet), k)
# 更新质心,直到变化量全为0
change, newCentroids = classify(dataSet, centroids, k)
while np.any(change != 0):
change, newCentroids = classify(dataSet, newCentroids, k)
centroids = sorted(newCentroids.tolist())
return centroids
# 高斯分布的概率密度函数
def prob(x, mu, sigma):
n = np.shape(x)[1]
expOn = float(-0.5 * (x - mu) * (np.linalg.inv(sigma)) * ((x - mu).T))
divBy = pow(2 * np.pi, n / 2) * pow(np.linalg.det(sigma), 0.5) # np.linalg.det 计算矩阵的行列式
return pow(np.e, expOn) / divBy
# EM算法
def EM(dataMat,centroids, maxIter,c):
m, n = np.shape(dataMat) # m=150, n=2
# 初始化参数
alpha = [1 / 3, 1 / 3, 1 / 3] # 系数
mu = np.mat(centroids) # 均值向量
# mu = random.sample(list(dataMat), c)
sigma = [np.mat([[0.1, 0], [0, 0.1]]) for x in range(3)] # 初始化协方差矩阵Σ
gamma = np.mat(np.zeros((m, c))) # γ(ik)
for i in range(maxIter):
for j in range(m):
sumAlphaMulP = 0
for k in range(c):
gamma[j, k] = alpha[k] * prob(dataMat[j, :], mu[k], sigma[k])
sumAlphaMulP += gamma[j, k]
for k in range(c):
gamma[j, k] /= sumAlphaMulP # 计算后验分布
sumGamma = np.sum(gamma, axis=0)
for k in range(c):
mu[k] = np.mat(np.zeros((1, n)))
sigma[k] = np.mat(np.zeros((n, n)))
for j in range(m):
mu[k] += gamma[j, k] * dataMat[j, :]
mu[k] /= sumGamma[0, k] # 更新均指向量
for j in range(m):
sigma[k] += gamma[j, k] * (dataMat[j, :] - mu[k]).T *(dataMat[j, :] - mu[k])
sigma[k] /= sumGamma[0, k] # 更新协方差矩阵
alpha[k] = sumGamma[0, k] / m # 更新混合系数
return gamma
# 高斯混合聚类
def gaussianCluster(dataMat,centroids,k):
m, n = np.shape(dataMat)
clusterAssign = np.mat(np.zeros((m, n)))
gamma = EM(dataMat,centroids,5,3)
centroids = np.array(centroids)
for i in range(m):
clusterAssign[i, :] = np.argmax(gamma[i, :]), np.amax(gamma[i, :])
for j in range(k):
pointsInCluster = dataMat[np.nonzero(clusterAssign[:, 0].A == j)[0]] # 将数据分类
centroids[j, :] = np.mean(pointsInCluster, axis=0) # 确定各均值中心,获得分类模型
return centroids, clusterAssign
def showCluster(dataMat, k, centroids, clusterAssment):
numSamples, dim = dataMat.shape
mark = ['o', '+', '*']
color = ['green','blue','orange']
for i in range(numSamples):
Index = int(clusterAssment[i, 0])
plt.plot(dataMat[i, 0], dataMat[i, 1],mark[Index],c = color[Index],markersize = 7)
for i in range(k):
plt.plot(centroids[i, 0], centroids[i, 1], marker='x',c = 'red',label = 'centroid',markersize = 10)
plt.title("GMM clustering results")
plt.xlabel("Sepal.Length")
plt.ylabel("Petal.Length")
plt.show()
if __name__=="__main__":
dataSet,label = loadDataSet() # 导入数据
dataMat = np.mat(dataSet)
centroids = kmeans(dataSet, 3) # 使用K_means算法初始化质心
centroids, clusterAssign = gaussianCluster(dataMat,centroids,3) # GMM算法
curr = 0
l = len(dataSet)
for i in range(l):
if label[i] == clusterAssign[i,0]:
curr += 1
currD = curr/l * 100
print("GMM准确率:",currD,"%")
showCluster(dataMat, 3, centroids, clusterAssign) # 可视化结果
三:实验结果

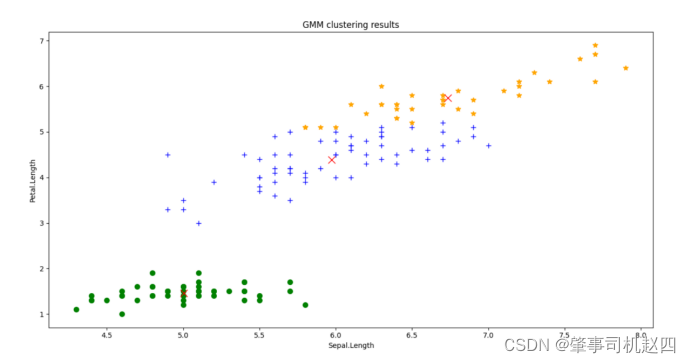
可以看到k-means聚类算法的聚类结果和高斯混合聚类算法在某些个体的分类上出现了差异,总的来说高斯混合聚类算法的分类准确度要高一些(这一点在第(4)小点进行讨论),但是能感觉到混合高斯聚类算法的收敛速度更慢,运行时间更长。
对于k-means聚类算法,必须要确定好生成簇的数目。当k取值过小时,簇内误方差会很大,训练出来的模型就会很差,但是当k过大时候又会导致分类过度,得不到想要的效果,因此k值的选取对于k-means算法来说至关重要。
参照图3,可以看到簇内误方差(SSE)图的拐点在k=3处取得,且k=3时其对应的轮廓系数(见图4)较大,而当k取4时,轮廓系数骤减。当轮廓系数为1时,说明这个点与周围簇距离较远,结果非常好,当它为0,说明这个点可能处在两个簇的边界上,当值为负时,暗含该点可能被误分了,所以k最合适的取值为3,正好符合鸢尾花的分类数量。