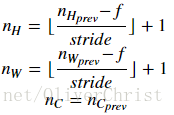构建卷积神经网络整体步骤
一、卷积
1.Zero padding
2.Convole window
3.Convolution forward
4.Convolution backward(Optional)
二、池化
1.Polling forward
2.create mask
3.distribute values
4.Pooling backward(Optional)

给矩阵X实施padding
def zero_pad(X, pad):
"""
Pad with zeros all images of the dataset X. The padding is applied to the height and width of an image,
as illustrated in Figure 1.
Argument:
X -- python numpy array of shape (m, n_H, n_W, n_C) representing a batch of m images
pad -- integer, amount of padding around each image on vertical and horizontal dimensions
X_pad=np.pad(X,((0,0),(pad,pad),(pad,pad),(0,0)),'constant')
构建卷积filter
代码实现
def conv_single_step(a_slice_prev, W, b):
Arguments:
a_slice_prev -- slice of input data of shape (f, f, n_C_prev),某一时刻filter覆盖的范围
W -- Weight parameters contained in a window - matrix of shape (f, f, n_C_prev)
b -- Bias parameters contained in a window - matrix of shape (1, 1, 1)
s = a_slice_prev * W
# Sum over all entries of the volume s.
Z = np.sum(s)
# Add bias b to Z. Cast b to a float() so that Z results in a scalar value.
Z = Z + float(b)
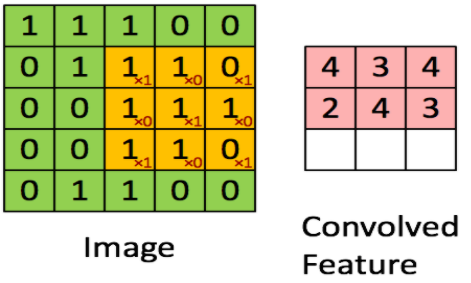
卷积层正向传播
代码实现
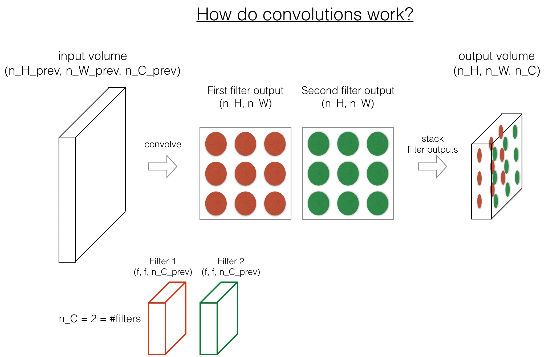
def conv_forward(A_prev, W, b, hparameters):
Arguments:
A_prev -- output activations of the previous layer, numpy array of shape (m, n_H_prev, n_W_prev, n_C_prev)
W -- Weights, numpy array of shape (f, f, n_C_prev, n_C)
b -- Biases, numpy array of shape (1, 1, 1, n_C)
hparameters -- python dictionary containing "stride" and "pad"
(m, n_H_prev, n_W_prev, n_C_prev) = A_prev.shape
# Retrieve dimensions from W's shape (≈1 line)
(f, f, n_C_prev, n_C) = W.shape
# Retrieve information from "hparameters" (≈2 lines)
stride = hparameters['stride']
pad = hparameters['pad']
# Compute the dimensions of the CONV output volume using the formula given above. Hint: use int() to floor. (≈2 lines)
n_H = int((n_H_prev-f+2*pad)/stride + 1)
n_W = int((n_W_prev-f+2*pad)/stride + 1)
# Initialize the output volume Z with zeros. (≈1 line)
Z = np.zeros((m,n_H,n_W,n_C))
A_prev_pad = zero_pad(A_prev,pad)
for i in range(m): # loop over the batch of training examples
a_prev_pad = A_prev_pad[i,:,:,:] # Select ith training example's padded activation
for h in range(n_H): # loop over vertical axis of the output volume
for w in range(n_W): # loop over horizontal axis of the output volume
for c in range(n_C): # loop over channels (= #filters) of the output volume
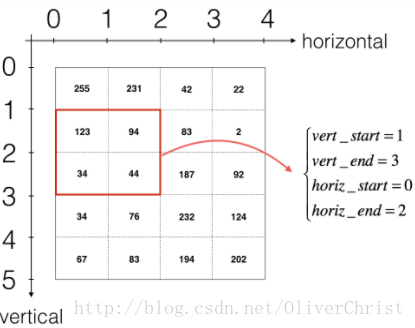
# Find the corners of the current "slice" (≈4 lines)
vert_start = stride*h
vert_end = vert_start + f
horiz_start = stride*w
horiz_end = horiz_start + f
# Use the corners to define the (3D) slice of a_prev_pad (See Hint above the cell). (≈1 line)
a_slice_prev = a_prev_pad[vert_start:vert_end,horiz_start:horiz_end,:]
# Convolve the (3D) slice with the correct filter W and bias b, to get back one output neuron. (≈1 line)
Z[i, h, w, c] = conv_single_step(a_slice_prev,W[:,:,:,c],b[:,:,:,c])
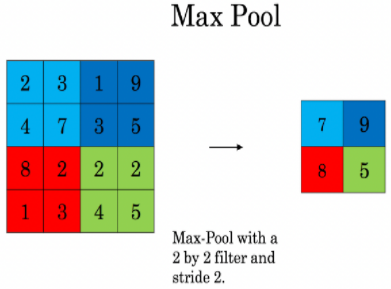
最大池化层前向传播
代码实现
def pool_forward(A_prev, hparameters, mode = "max"):
Arguments:
A_prev -- Input data, numpy array of shape (m, n_H_prev, n_W_prev, n_C_prev)
hparameters -- python dictionary containing "f" and "stride"
mode -- the pooling mode you would like to use, defined as a string ("max" or "average")
# Retrieve dimensions from the input shape
(m, n_H_prev, n_W_prev, n_C_prev) = A_prev.shape
# Retrieve hyperparameters from "hparameters"
f = hparameters["f"]
stride = hparameters["stride"]
# Define the dimensions of the output
n_H = int(1 + (n_H_prev - f) / stride)
n_W = int(1 + (n_W_prev - f) / stride)
n_C = n_C_prev
# Initialize output matrix A
A = np.zeros((m, n_H, n_W, n_C))
### START CODE HERE ###
for i in range(m): # loop over the training examples
for h in range(n_H): # loop on the vertical axis of the output volume
for w in range(n_W): # loop on the horizontal axis of the output volume
for c in range (n_C): # loop over the channels of the output volume
# Find the corners of the current "slice" (≈4 lines)
vert_start = h*stride
vert_end = vert_start + f
horiz_start = w*stride
horiz_end = horiz_start + f
# Use the corners to define the current slice on the ith training example of A_prev, channel c. (≈1 line)
a_prev_slice = A_prev[vert_start:vert_end,horiz_start:horiz_end,c]
# Compute the pooling operation on the slice. Use an if statment to differentiate the modes. Use np.max/np.mean.
A[i, h, w, c] = np.max(a_prev_slice)
参考Andrew Ng深度学习课程。