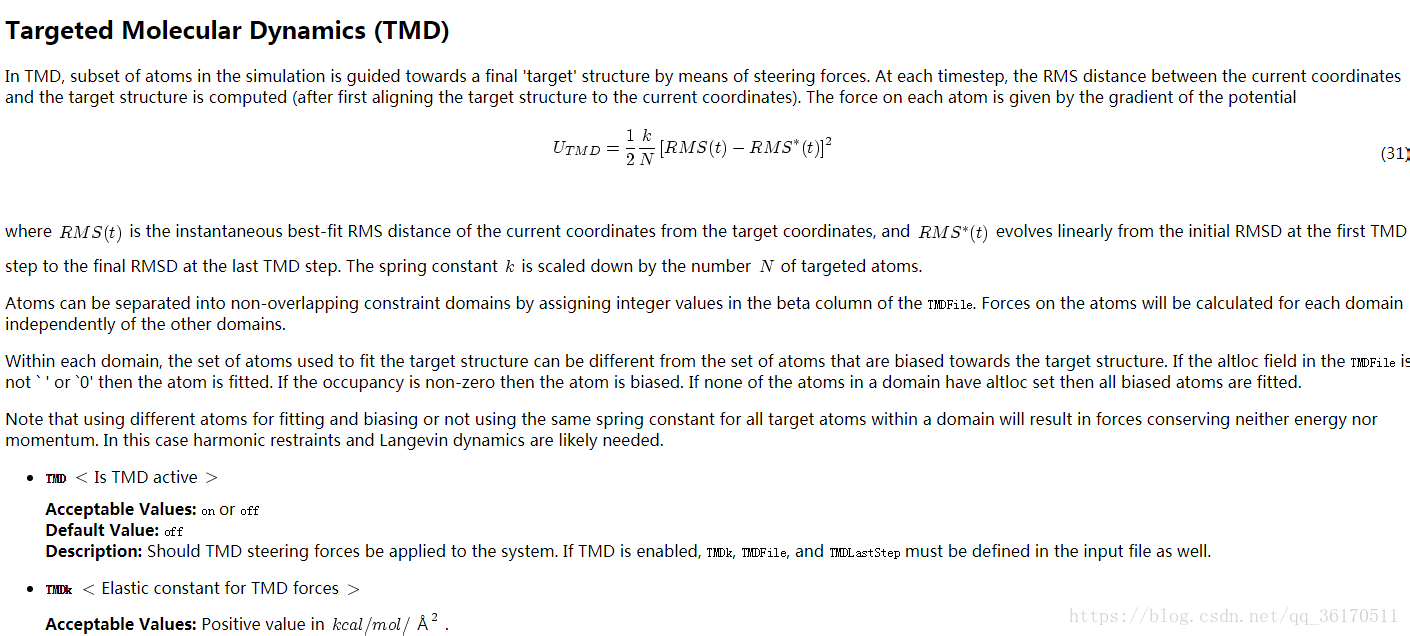
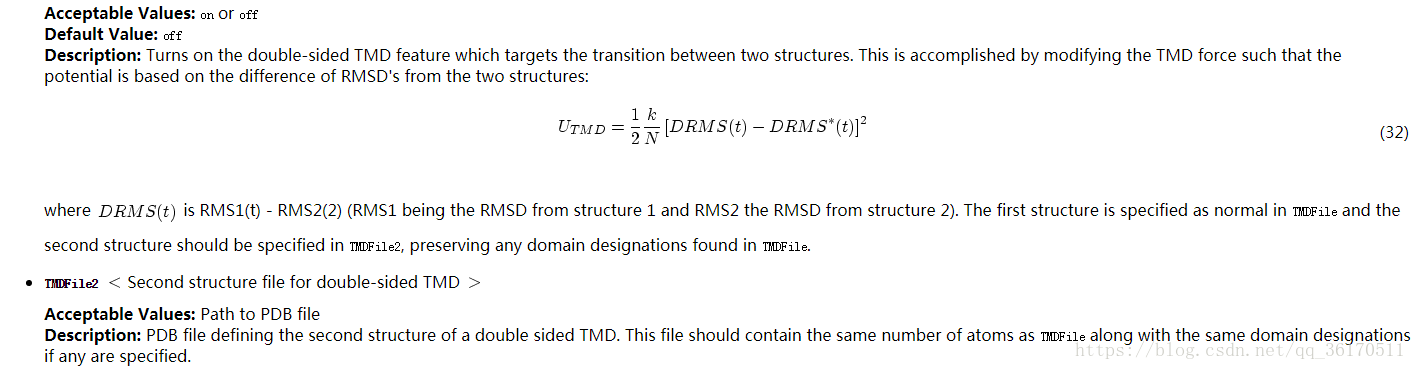
下面附上靶向分子动力学的conf文件,具体问题具体分析,参数根据自己体系进行修改。TMD比常规MD的conf不同就在于EXTRA PARAMETERS 这部分多了一些参数。具体设置可参考上述TMD介绍。
conf:
#############################################################
## JOB DESCRIPTION ##
#############################################################
# Min. and Eq. of KcsA
# embedded in POPC membrane, ions and water.
# Protein constrained. PME, Constant Pressure.
#############################################################
## ADJUSTABLE PARAMETERS ##
#############################################################
structure p-gpoutward_popc_water_ion.psf
coordinates p-gpoutward_popc_water_ion.pdb
outputName p-gpoutward_popc_water_ion_tmd
set temperature 310
# Continuing a job from the restart files
if {1} {
set inputname p-gpoutward_popc_water_ion_eqm2
binCoordinates $inputname.restart.coor
binVelocities $inputname.restart.vel ;# remove the "temperature" entry if you use this!
extendedSystem $inputname.restart.xsc
}
firsttimestep 1775000
#############################################################
## SIMULATION PARAMETERS ##
#############################################################
# Input
paraTypeCharmm on
parameters par_all27_prot_lipidNBFIX.prm
# NOTE: Do not set the initial velocity temperature if you
# have also specified a .vel restart file!
# temperature $temperature
# Periodic Boundary Conditions
# NOTE: Do not set the periodic cell basis if you have also
# specified an .xsc restart file!
if {0} {
cellBasisVector1 164. 0. 0.
cellBasisVector2 0. 167. 0.
cellBasisVector3 0. 0. 180.
cellOrigin -0.40401795506477356 -1.9578770399093628 7.172184467315674
}
wrapWater on
wrapAll on
# Force-Field Parameters
exclude scaled1-4
1-4scaling 1.0
cutoff 12.
switching on
switchdist 10.
pairlistdist 13.5
# Integrator Parameters
timestep 1.0 ;# 2fs/step
# rigidBonds all ;# needed for 2fs steps
nonbondedFreq 1
fullElectFrequency 2
stepspercycle 20
#PME (for full-system periodic electrostatics)
if {1} {
PME yes
PMEGridSizeX 145
PMEGridSizeY 145
PMEGridSizeZ 183
}
# Constant Temperature Control
langevin on ;# do langevin dynamics
langevinDamping 1 ;# damping coefficient (gamma) of 5/ps
langevinTemp $temperature
# Constant Pressure Control (variable volume)
if {1} {
useGroupPressure yes ;# needed for 2fs steps
useFlexibleCell yes ;# no for water box, yes for membrane
useConstantArea yes ;# no for water box, yes for membrane
langevinPiston on
langevinPistonTarget 1.01325 ;# in bar -> 1 atm
langevinPistonPeriod 400.
langevinPistonDecay 200.
langevinPistonTemp $temperature
}
restartfreq 500 ;# 1000steps = every 2ps
dcdfreq 500
xstFreq 500
outputEnergies 500
outputPressure 500
# Fixed Atoms Constraint (set PDB beta-column to 1)
if {0} {
fixedAtoms on
fixedAtomsFile nottails.fix.pdb
fixedAtomsCol B
fixedAtomsForces on
}
#############################################################
## EXTRA PARAMETERS ##
#############################################################
# Put here any custom parameters that are specific to
# this job (e.g., SMD, TclForces, etc...)
# constraints on
# consexp 2
# consref p-gpoutward_popc_water_ion.pdb
# conskfile p-gpoutward_popc_water_ion_ca.pdb
# conskcol B
# margin 3
tclforces on
set waterCheckFreq 100
set lipidCheckFreq 100
set allatompdb p-gpoutward_popc_water_ion.pdb
tclForcesScript keep_water_out.tcl
# eFieldOn yes
# eField 0 0 -0.155
TMD on
TMDFile p-gpinward_tmd_nbd.pdb
TMDk 515
TMDOutputFreq 500
TMDFinalRMSD 0
TMDFirstStep 1775000
TMDLastStep 6775000
#############################################################
## EXECUTION SCRIPT ##
#############################################################
# Minimization
if {0} {
minimize 20000
reinitvels $temperature
}
run 5000000 ;# 5 ns