本文来源于各路集合:https://blog.csdn.net/justbeherhero/category_12041739.html
思考: https://mapengsen.blog.csdn.net/article/details/128617817
识别原子和骨架的位置和类型,然后组装
一、2022-BIB | ABC-Net: a divide-and-conquer based deep learning architecture for SMILES recognition from molecular image
ABC-Net:基于分治法的分子ImageToSMILES的DL架构
一些解析:BIB |基于分而治之的分子图片识别深度学习框架 - 知乎
https://blog.csdn.net/justBeHerHero/article/details/127322013
ABC-Net:从分子图像中进行SMILES识别的深度学习框架 - 墨天轮
论文: https://arxiv.org/abs/2002.10200
code :GitHub - zhang-xuan1314/ABC-Net: ABC-Net for molecular image recognition

二、2021-BNAIC/Benelearn | Self-labeling of Fully Mediating Representations by Graph Alignment
paper:Self-labeling of Fully Mediating Representations by Graph Alignment | SpringerLink
code : GitHub - biolearning-stadius/chemgrapher-self-rich-labeling
三、2022-Briefings in bioinformatics | Multi-modal chemical information reconstruction from images and texts for exploring the near-drug space
基于images-texts的Multi-modal结构信息重建以探索near-drug空间
https://blog.csdn.net/justBeHerHero/article/details/127397117
四、2020-JCIM-ChemGrapher: Optical Graph Recognition of Chemical Compounds by Deep Learning
https://pubs.acs.org/doi/10.1021/acs.jcim.0c00459
https://blog.csdn.net/justBeHerHero/article/details/127223664
code:https://github.com/biolearning-stadius/chemgrapher-self-rich-labeling
Encoder-Decoder:
一、2021-Chemical Science | Img2Mol:accurate SMILES recognition from molecular graphical depictions
https://blog.csdn.net/justBeHerHero/article/details/127175730
code: GitHub - bayer-science-for-a-better-life/Img2Mol
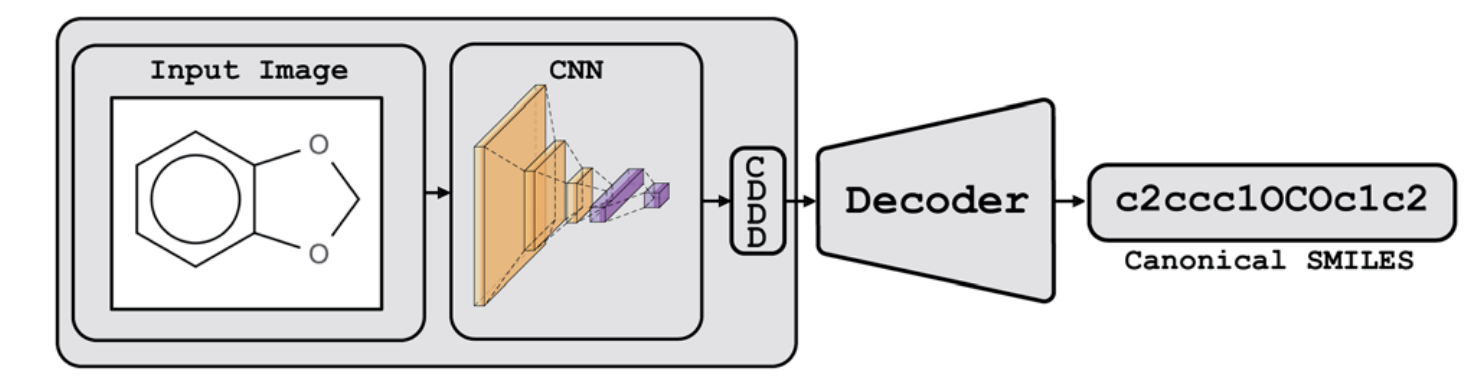
这个论文很简单,就是将image 通过CNN encoder ,然后通过CDDD层【把将SMILES标准化的模型中的512宽度的layer叫做CDDD】,再通过Decoder 将CNN encoder的信息成SMILES
与ABC-Net这种“ 拆分-识别-组装 ”的处理方式完全不同
二、2022_Robust Molecular Image Recognition: A Graph Generation Approach
paper:https://arxiv.org/pdf/2205.14311.pdf
https://blog.csdn.net/justBeHerHero/article/details/127238332
三、2022-Bioinformatics | MICER:A Pre-trained Encoder-Decoder Architecture for Molecular Image Captioning
https://blog.csdn.net/justBeHerHero/article/details/127244027
Transformer:
一、2021-Cheminform | DECIMER 1.0: deep learning for chemical image recognition using transformers
https://blog.csdn.net/justBeHerHero/article/details/127188337
code:The DECIMER 2.0:https://github.com/Kohulan/DECIMER-Image_Transformer
二、2021-Chemistry Methods | Image2SMILES: Transformer-Based Molecular Optical Recognition Engine
https://blog.csdn.net/justBeHerHero/article/details/127187994
https://chemistry-europe.onlinelibrary.wiley.com/doi/full/10.1002/cmtd.202100069
三、2022-Cheminform | SwinOCSR: end-to-end optical chemical structure recognition using a Swin Transformer
SwinOCSR+:基于Swin Transformer的end-to-end光学化学结构识别
paper : SwinOCSR: end-to-end optical chemical structure recognition using a Swin Transformer | Journal of Cheminformatics | Full Text
code: GitHub - suanfaxiaohuo/SwinOCSR
https://blog.csdn.net/justBeHerHero/article/details/127487296
四、2022-ICASSP | Image-to-Graph Transformers for Chemical Structure Recognition
杂项
2021-Chemical Science【手写识别】 | ChemPix: automated recognition of hand-drawn hydrocarbon structures using deep learning
https://blog.csdn.net/justBeHerHero/article/details/127191454
code: https://github.com/mtzgroup/ChemPixCH
2022-Journal of Cheminformatics【手写识别】丨DECIMER—hand-drawn molecule images dataset
DECIMER—hand-drawn molecule images dataset | Journal of Cheminformatics | Full Text
2022-JCIM-MolMiner: You only look once for chemical structure recognition
paper:https://arxiv.org/pdf/2205.11016.pdf
2019-JCIM 【只是用了深度神经网络】| Molecular Structure Extraction From Documents Using Deep Learning
https://blog.csdn.net/justBeHerHero/article/details/127187714
https://pubs.acs.org/doi/10.1021/acs.jcim.8b00669
识别软件 InDraw 和 Mathpix OCR:
免费的InDraw:
上海鹰谷公司于2018年5月公布了其结构式智能识别的专利,这篇长达12页的专利比较详细地介绍了对含有结构式图片的处理和提取方法。:【一种化学结构式的智能识别方法】
附:
我自己下载了一个,然后识别了一下,识别的真不行,果断卸载了