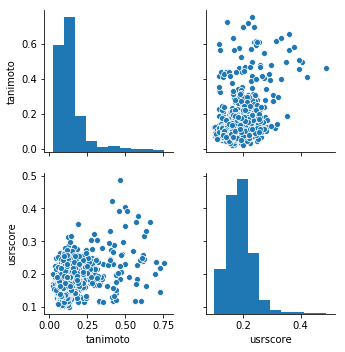
USRCAT
USRCAT是基于形状的方法,它的工作速度非常快。代码是免费提供的,如果要使用代码,用户需要安装它。
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3505738/
RDKit代码:
import os
import seaborn as sns
import pandas as pd
from rdkit import Chem
from rdkit.Chem import rdBase
from rdkit.Chem import RDConfig
from rdkit.Chem import AllChem
from rdkit.Chem.rdMolDescriptors import GetUSRScore, GetUSRCAT
from rdkit.Chem import DataStructs
print( rdBase.rdkitVersion )
mols = [ mol for mol in Chem.SDMolSupplier( "cdk2.sdf" ) ]
for mol in mols:
AllChem.EmbedMolecule( mol,
useExpTorsionAnglePrefs = True,
useBasicKnowledge = True )
usrcats = [ GetUSRCAT( mol ) for mol in mols ]
fps = [ AllChem.GetMorganFingerprintAsBitVect( mol, 2 ) for mol in mols ]
data = { "tanimoto":[], "usrscore":[] }
for i in range( len( usrcats )):
for j in range( i ):
tc = DataStructs.TanimotoSimilarity( fps[ i ], fps[ j ] )
score = GetUSRScore( usrcats[ i ], usrcats[ j ] )
data["tanimoto"].append( tc )
data["usrscore"].append( score )
print( score, tc )
df = pd.DataFrame( data )
fig = sns.pairplot( df )
fig.savefig( 'plot.png' )