Pytorch与QSAR
尝试使用pytorch和RDKit构建QSAR模型
环境依赖
pip install pprint
pip install argparse
#安装rdkit
conda install -c rdkit rdkit基于Pytorch和RDKit的QSAR模型代码:
#!usr/bin/python3
import pprint
import argparse
import torch
import torch.optim as optim
from torch import nn as nn
import torch.nn.functional as F
from torch.autograd import Variable
from rdkit import Chem
from rdkit.Chem import AllChem
from rdkit.Chem import DataStructs
import numpy as np
#from sklearn import preprocessing
def base_parser():
parser = argparse.ArgumentParser("This is simple test of pytorch")
parser.add_argument("trainset", help="sdf for train")
parser.add_argument("testset", help="sdf for test")
parser.add_argument("--epochs", default=150)
return parser
parser = base_parser()
args = parser.parse_args()
traindata = [mol for mol in Chem.SDMolSupplier(args.trainset) if mol is not None]
testdata = [mol for mol in Chem.SDMolSupplier(args.testset) if mol is not None]
def molsfeaturizer(mols):
fps = []
for mol in mols:
arr = np.zeros((0,))
fp = AllChem.GetMorganFingerprintAsBitVect(mol, 2)
DataStructs.ConvertToNumpyArray(fp, arr)
fps.append(arr)
fps = np.array(fps, dtype = np.float)
return fps
classes = {"(A) low":0, "(B) medium":1, "(C) high":2}
#classes = {"(A) low":0, "(B) medium":1, "(C) high":1}
trainx = molsfeaturizer(traindata)
testx = molsfeaturizer(testdata)
# for pytorch, y must be long type!!
trainy = np.array([classes[mol.GetProp("SOL_classification")] for mol in traindata], dtype=np.int64)
testy = np.array([classes[mol.GetProp("SOL_classification")] for mol in testdata], dtype=np.int64)
#在pytorch中构建模型,定义每个层和整个结构
X_train = torch.from_numpy(trainx)
X_test = torch.from_numpy(testx)
Y_train = torch.from_numpy(trainy)
Y_test = torch.from_numpy(testy)
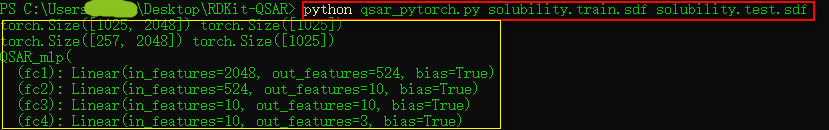
print(X_train.size(),Y_train.size())
print(X_test.size(), Y_train.size())
class QSAR_mlp(nn.Module):
def __init__(self):
super(QSAR_mlp, self).__init__()
self.fc1 = nn.Linear(2048, 524)
self.fc2 = nn.Linear(524, 10)
self.fc3 = nn.Linear(10, 10)
self.fc4 = nn.Linear(10,3)
def forward(self, x):
x = x.view(-1, 2048)
h1 = F.relu(self.fc1(x))
h2 = F.relu(self.fc2(h1))
h3 = F.relu(self.fc3(h2))
output = F.sigmoid(self.fc4(h3))
return output
#构建训练和预测模型
model = QSAR_mlp()
print(model)
losses = []
optimizer = optim.Adam( model.parameters(), lr=0.005)
for epoch in range(args.epochs):
data, target = Variable(X_train).float(), Variable(Y_train).long()
optimizer.zero_grad()
y_pred = model(data)
loss = F.cross_entropy(y_pred, target)
print("Loss: {}".format(loss.data[0]))
loss.backward()
optimizer.step()
pred_y = model(Variable(X_test).float())
predicted = torch.max(pred_y, 1)[1]
for i in range(len(predicted)):
print("pred:{}, target:{}".format(predicted.data[i], Y_test[i]))
print( "Accuracy: {}".format(sum(p==t for p,t in zip(predicted.data, Y_test))/len(Y_test)))测试模型
python qsar_pytorch.py solubility.train.sdf solubility.test.sdf留一个公众号二维码,欢迎大家一起交流学习!