欢迎关注我的CSDN:https://spike.blog.csdn.net/
本文地址:https://blog.csdn.net/caroline_wendy/article/details/131403263
论文:ProteinChat: Towards Enabling ChatGPT-Like Capabilities on Protein 3D Structures
工程:https://github.com/UCSD-AI4H/proteinchat
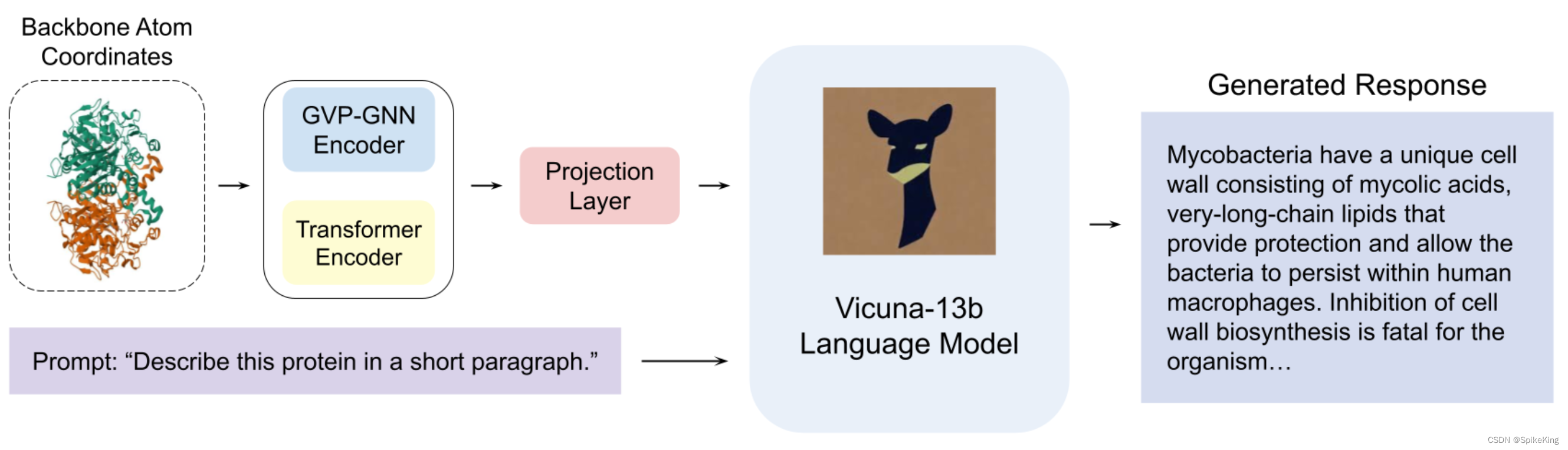
ProteinChat 是基于大型语言模型(LLM)的原型系统,能够根据蛋白质的三维结构进行问答和文本解释。ProteinChat 利用一个复合编码器块和一个 LLM 解码器块,协同工作,提供蛋白质相关的洞察。复合编码器块结合了一个图神经网络(GNN)编码器块和一个 Transformer 编码器块,有效地从蛋白质结构中提取重要特征。LLM 解码器利用编码器块生成的蛋白质嵌入和用户的问题,生成信息丰富的答案。为了训练 ProteinChat,构建了RCSB-PDB蛋白质描述数据集,包含了143,508个来自公开可用资源的蛋白质-描述对。ProteinChat 是第一个利用LLM来研究蛋白质的工作,为进一步探索和利用ChatGPT-like系统在蛋白质研究中的应用奠定了基础。
参考:LLM - 搭建 DrugGPT 结合药物化学分子知识的 ChatGPT 系统
1. 配置环境
下载工程与配置 conda 环境:
# 文件较多,下载需要一段时间
# git clone https://github.com/UCSD-AI4H/proteinchat
git clone [email protected]:UCSD-AI4H/proteinchat.git # 建议使用git模式下载
cd proteinchat
conda env create -f environment.yml
conda activate proteinchat
pip install einops
建议参考 DrugGPT 的配置方案。
安装 pytorch 相关的包:
nvidia-smi
conda install pytorch==1.12.1 torchvision==0.13.1 torchaudio==0.12.1 cudatoolkit=11.6 -c pytorch -c conda-forge
验证 PyTorch 通过:
python -c "import torchvision; print(torchvision.__version__)"
0.13.1
python -c "import torch; print(torch.__version__)"
1.12.1
准备数据集:
# 链接
https://drive.google.com/u/0/uc?id=1xdiBP-FPMfwpMGBUPAKd0FyRrqQtxAEK&export=download # qa_all.json (499M)
https://drive.google.com/u/0/uc?id=1iMgPyiIzpvXdKiNsXnRKn2YpmP92Xyub&export=download # abstract.json (182M)
https://drive.google.com/u/0/uc?id=1AeJW5BY5C-d8mKJjAULTax6WA4hzWS0N&export=download # 暂时无法访问
pip install gdown
gdown https://drive.google.com/uc?id=1xdiBP-FPMfwpMGBUPAKd0FyRrqQtxAEK # 已下载完成
gdown https://drive.google.com/uc?id=1iMgPyiIzpvXdKiNsXnRKn2YpmP92Xyub # 已下载完成
下载 Google云盘使用 gdown 软件,参考:GitHub gdown,注意本地可以使用,服务器需要连接外网。
ESM-IF1 数据问题,等待解决,暂时使用临时数据。
2. 训练模型
准备辅助模型:
- bert-base-uncased:参考 CSDN - Hugging Face 工程 BERT base model (uncased) 配置
- vicuna-13b-weight:参考 CSDN - 基于 Vicuna-13B 参数计算搭建私有 ChatGPT 在线聊天
准备训练数据:
pt:已经从 ESM-IF1 提取的 PDB 特征。ann.json:PDB的描述信息
其中,ann.json 的数据如下:
[
{
"pdb_id": "6nk3",
"caption": "Mxra8 is a receptor for multiple arthritogenic alphaviruses that cause debilitating acute and chronic musculoskeletal disease in humans. Herein, we present a 2.2\u00a0\u00c5 resolution X-ray crystal structure of Mxra8 and 4 to 5\u00a0\u00c5 resolution cryo-electron microscopy reconstructions of Mxra8 bound to chikungunya (CHIKV) virus-like particles and infectious virus. The Mxra8 ectodomain contains two strand-swapped Ig-like domains oriented in a unique disulfide-linked head-to-head arrangement. Mxra8 binds by wedging into a cleft created by two adjacent CHIKV E2-E1 heterodimers in one trimeric spike and engaging a neighboring spike. Two binding modes are observed with the fully mature VLP, with one Mxra8 binding with unique contacts. Only the high-affinity binding mode was observed in the complex with infectious CHIKV, as viral maturation and E3 occupancy appear to influence receptor binding-site usage. Our studies provide insight into how Mxra8 binds CHIKV and creates a path for developing alphavirus entry inhibitors."
},
{
"pdb_id": "6dbp",
"caption": "The MUSASHI (MSI) family of RNA binding proteins (MSI1 and MSI2) contribute to a wide spectrum of cancers including acute myeloid leukemia. We find that the small molecule Ro 08-2750 (Ro) binds directly and selectively to MSI2 and competes for its RNA binding in biochemical assays. Ro treatment in mouse and human myeloid leukemia cells results in an increase in differentiation and apoptosis, inhibition of known MSI-targets, and a shared global gene expression signature similar to shRNA depletion of MSI2. Ro demonstrates in vivo inhibition of c-MYC and reduces disease burden in a murine AML leukemia model. Thus, we identify a small molecule that targets MSI's oncogenic activity. Our study provides a framework for targeting RNA binding proteins in cancer."
},
...
]
修改训练脚本train_esm.py,支持使用 mini 训练集 或 全量数据集:
datasets_raw = ESMDataset(pdb_root="data/esm_subset/pt",
ann_paths="data/esm_subset/ann.json",
chain="A")
修改配置文件 minigpt4/configs/models/minigpt4.yaml:
llama_model: "workspace/vicuna-13b-weight"
修改模型文件 minigpt4/models/blip2.py,即辅助模型 bert-base-uncased 的路径,即:
class Blip2Base(BaseModel):
@classmethod
def init_tokenizer(cls):
# tokenizer = BertTokenizer.from_pretrained("bert-base-uncased")
tokenizer = BertTokenizer.from_pretrained("workspace_v2/bert-base-uncased")
tokenizer.add_special_tokens({
"bos_token": "[DEC]"})
return tokenizer
运行训练脚本:
nvidia-smi
CUDA_VISIBLE_DEVICES=2 bash finetune.sh
训练日志,如下:
2023-06-26 17:37:16,309 [INFO] Start training
2023-06-26 17:37:16,317 [INFO] Start training epoch 15, 762 iters per inner epoch.
Train: data epoch: [15] [ 0/762] eta: 0:33:20 lr: 0.000010 loss: 1.3494 time: 2.6249 data: 0.0000 max mem: 35719
Train: data epoch: [15] [ 50/762] eta: 0:03:14 lr: 0.000010 loss: 1.3266 time: 0.2184 data: 0.0000 max mem: 35719
...
Train: data epoch: [99] [750/762] eta: 0:00:02 lr: 0.000001 loss: 0.5560 time: 0.1983 data: 0.0000 max mem: 35720
Train: data epoch: [99] [761/762] eta: 0:00:00 lr: 0.000001 loss: 0.2065 time: 0.2299 data: 0.0000 max mem: 35720
Train: data epoch: [99] Total time: 0:02:52 (0.2260 s / it)
2023-06-26 21:41:19,045 [INFO] Averaged stats: lr: 0.0000 loss: 0.4527
2023-06-26 21:41:19,132 [INFO] No validation splits found.
2023-06-26 21:41:19,139 [INFO] Saving checkpoint at epoch 99 to proteinchat/minigpt4/output/minigpt4_stage2_esm/20230626164/checkpoint_99.pth.
2023-06-26 21:41:19,377 [INFO] No validation splits found.
2023-06-26 21:41:19,377 [INFO] Training time 4:47:07
Bug: RuntimeError: The server socket has failed to listen on any local network address. The server socket has failed to bind to [::]:29500 (errno: 98 - Address already in use). The server socket has failed to bind to 0.0.0.0:29500 (errno: 98 - Address already in use).
参考:CSDN - Pytorch中DDP :The server socket has failed to bind to [::]:29500
具体而言,修改 finetune.sh 文件,torchrun 增加参数 --master_port,将默认的29500修改为29501,即可:
torchrun --master_port=29501 train_esm.py --cfg-path train_configs/minigpt4_stage2_esm.yaml
3. 推理模型
已训练完成,复制模型:
cp minigpt4/output/minigpt4_stage2_esm/20230626164/checkpoint_99.pth ckpt/checkpoint_99.pth
模型位于 ckpt/checkpoint_99.pth。
修改推理配置 eval_configs/proteinchat_eval.yaml
ckpt: 'ckpt/checkpoint_99.pth'
修改网页脚本 demo_esm.py,增加端口和链接:
# 默认是 127.0.0.1 无法访问
demo.launch(share=True, enable_queue=True, server_name="0.0.0.0", server_port=9300)
运行程序:
CUDA_VISIBLE_DEVICES=2 bash demo.sh
运行成功,推理特征速度较慢。