Model Selection & Evaluation
Agenda
- Cross Validation
- Hyperparameter Tuning
- Model Evaluation
- Model Persistance
- Validation Curves
- Learning Curves
- 交叉验证
- 超参数调整
- 模型评估
- 模型的持久性
- 验证曲线
- 学习曲线
Cross Validation交叉验证
- Simple models underfit.
- Accuracy for training data & validation data is not much different.
- But, accurcy ain’t that great.
- This situation is of low variance & high bias
- On moving towards complex models, accuracy improves.
- But, gap between accuracy on training data & validation data increases
- This situation is of high variance & low bias
- 简单模型的拟合度不足。
- 训练数据和验证数据的准确度没有太大区别。
- 但是,准确度没有那么高。
- 这种情况是低方差和高偏差。
- 在向复杂模型发展的过程中,准确度有所提高。
- 但是,训练数据和验证数据的准确性之间的差距会增加。
- 这种情况是高偏差和低偏差。

- We need to compare across models to find the best model.
- We need to compare across all hyper-parameters for a particular model.
- The data that is used for training should not be used for validation.
- The validation accuracy is the one that we claims
- 我们需要在不同模型之间进行比较,以找到最佳模型。
- 我们需要对一个特定模型的所有超参数进行比较。
- 用于训练的数据不应该被用于验证。
- 验证精度就是我们所声称的验证精度。
from sklearn.tree import DecisionTreeClassifier
from sklearn.datasets import load_digits
import matplotlib.pyplot as plt
%matplotlib inline
digits = load_digits()
plt.imshow(digits.images[0],cmap='gray')

from sklearn.model_selection import train_test_split
dt = DecisionTreeClassifier(max_depth=10)
trainX, testX, trainY, testY = train_test_split(digits.data, digits.target)
dt.fit(trainX,trainY)
DecisionTreeClassifier(class_weight=None, criterion='gini', max_depth=10,
max_features=None, max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, presort=False,
random_state=None, splitter='best')
dt.score(testX,testY)
0.8355555555555556
dt.score(trainX,trainY)
0.9740163325909429
- Decreasing the complexity of model
- 降低模型的复杂性
dt = DecisionTreeClassifier(max_depth=7)
dt.fit(trainX,trainY)
DecisionTreeClassifier(class_weight=None, criterion='gini', max_depth=7,
max_features=None, max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, presort=False,
random_state=None, splitter='best')
dt.score(testX,testY)
0.8155555555555556
dt.score(trainX,trainY)
0.8864142538975501
- Observation : With decrease in complexity the gap in training & validation accuracy also decreased
- 观察:随着复杂度的降低,训练和验证精度的差距也在缩小。
Cross Validation API交叉验证API
- Splits data into k parts.
- Use k - 1 parts for training the model
- Use kth part for validation
- Repeat the above steps multiple times to get a generalized behaviour
- 将数据分割成k个部分。
- 使用k-1部分来训练模型。
- 使用第K部分进行验证
- 重复上述步骤多次,得到一个通用的行为。
from sklearn.model_selection import cross_val_score
scores = cross_val_score(dt, digits.data, digits.target)
d:\Anaconda3\lib\site-packages\sklearn\model_selection\_split.py:1978: FutureWarning: The default value of cv will change from 3 to 5 in version 0.22. Specify it explicitly to silence this warning.
warnings.warn(CV_WARNING, FutureWarning)
scores
array([0.68604651, 0.8096828 , 0.74161074])
scores.mean()
0.7457800181857993
Cross-validate Function : Scores for multiple matrices交叉验证函数:多个矩阵的分数
from sklearn.model_selection import cross_validate
scoring = ['precision_macro', 'recall_macro', 'accuracy']
results=cross_validate(dt, digits.data, digits.target, scoring=scoring, cv=5)
results
{'fit_time': array([0.01800108, 0.01200056, 0.01400089, 0.01300073, 0.01400089]),
'score_time': array([0.00300026, 0.00300002, 0.00200009, 0.00300002, 0.00300002]),
'test_precision_macro': array([0.7732771 , 0.71087424, 0.77524663, 0.78964348, 0.7585891 ]),
'test_recall_macro': array([0.76278636, 0.66593093, 0.76876662, 0.77198413, 0.74553688]),
'test_accuracy': array([0.76373626, 0.66574586, 0.76880223, 0.77310924, 0.74366197])}
for k, v in results.items():
print(k,end=' ')
print(v)
fit_time [0.01800108 0.01200056 0.01400089 0.01300073 0.01400089]
score_time [0.00300026 0.00300002 0.00200009 0.00300002 0.00300002]
test_precision_macro [0.7732771 0.71087424 0.77524663 0.78964348 0.7585891 ]
test_recall_macro [0.76278636 0.66593093 0.76876662 0.77198413 0.74553688]
test_accuracy [0.76373626 0.66574586 0.76880223 0.77310924 0.74366197]
results.keys()
dict_keys(['fit_time', 'score_time', 'test_precision_macro', 'test_recall_macro', 'test_accuracy'])
import pandas as pd
results=pd.DataFrame(results.values(),index=results.keys()).T
results
| fit_time | score_time | test_precision_macro | test_recall_macro | test_accuracy | |
|---|---|---|---|---|---|
| 0 | 0.018001 | 0.003 | 0.773277 | 0.762786 | 0.763736 |
| 1 | 0.012001 | 0.003 | 0.710874 | 0.665931 | 0.665746 |
| 2 | 0.014001 | 0.002 | 0.775247 | 0.768767 | 0.768802 |
| 3 | 0.013001 | 0.003 | 0.789643 | 0.771984 | 0.773109 |
| 4 | 0.014001 | 0.003 | 0.758589 | 0.745537 | 0.743662 |
Stratification for dealing with imbalanced Classes分层处理不平衡类的问题
- StratifiedKFold
- Class frequencies are preserved in data splitting
- 分层KFold
- 类频率在数据拆分中得到保留
import numpy as np
Y = np.append(np.ones(12),np.zeros(6))
X = np.ones((18,3))
from sklearn.model_selection import StratifiedKFold
skf = StratifiedKFold(n_splits=3)
list(skf.split(X,Y))
[(array([ 4, 5, 6, 7, 8, 9, 10, 11, 14, 15, 16, 17]),
array([ 0, 1, 2, 3, 12, 13])),
(array([ 0, 1, 2, 3, 8, 9, 10, 11, 12, 13, 16, 17]),
array([ 4, 5, 6, 7, 14, 15])),
(array([ 0, 1, 2, 3, 4, 5, 6, 7, 12, 13, 14, 15]),
array([ 8, 9, 10, 11, 16, 17]))]
Y[[ 4, 5, 6, 7, 8, 9, 10, 11, 14, 15, 16, 17]]
array([1., 1., 1., 1., 1., 1., 1., 1., 0., 0., 0., 0.])
Hyperparameter Tuning 超参数调整
- Model parameters are learnt by learning algorithms based on data
- Hyper-parameters needs to be configured
- Hyper-parameters are data dependent & many times need experiments to find the best
- sklearn provides GridSerach for finding the best hyper-parameters
Exhaustive GridSearch穷尽的网格搜索
-
Searches sequentially for all the configued params
-
For all possible combinations
-
模型参数由学习算法根据数据学习算法来学习。
-
需要配置超参数。
-
超参数依赖于数据,很多时候需要通过实验来找到最佳的参数。
-
sklearn提供了GridSerach,用于寻找最佳的超参数。
-
按顺序搜索所有配置的参数。
-
对于所有可能的组合
trainX, testX, trainY, testY = train_test_split(digits.data, digits.target)
dt = DecisionTreeClassifier()
from sklearn.model_selection import GridSearchCV
grid_search = GridSearchCV(dt, param_grid={'max_depth':range(5,30,5)}, cv=5)
grid_search.fit(digits.data,digits.target)
d:\Anaconda3\lib\site-packages\sklearn\model_selection\_search.py:814: DeprecationWarning: The default of the `iid` parameter will change from True to False in version 0.22 and will be removed in 0.24. This will change numeric results when test-set sizes are unequal.
DeprecationWarning)
GridSearchCV(cv=5, error_score='raise-deprecating',
estimator=DecisionTreeClassifier(class_weight=None,
criterion='gini', max_depth=None,
max_features=None,
max_leaf_nodes=None,
min_impurity_decrease=0.0,
min_impurity_split=None,
min_samples_leaf=1,
min_samples_split=2,
min_weight_fraction_leaf=0.0,
presort=False, random_state=None,
splitter='best'),
iid='warn', n_jobs=None, param_grid={'max_depth': range(5, 30, 5)},
pre_dispatch='2*n_jobs', refit=True, return_train_score=False,
scoring=None, verbose=0)
grid_search.best_params_
{'max_depth': 20}
grid_search.best_score_
0.7868670005564831
grid_search.best_estimator_
DecisionTreeClassifier(class_weight=None, criterion='gini', max_depth=20,
max_features=None, max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, presort=False,
random_state=None, splitter='best')
RandomizedSearch随机搜索
- Unlike GridSearch, not all parameters are tried & tested
- But rather a fixed number of parameter settings is sampled from the specified distributions.
Comparing GridSearch and RandomSearchCV
- 与GridSearch不同,不是所有的参数都是经过测试的。
- 而是从指定的分布中抽出一个固定数量的参数设置。
比较GridSearch和RandomSearchCV。
from time import time
#randint is an intertor for generating numbers between range specified
from scipy.stats import randint
X = digits.data
Y = digits.target
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import RandomizedSearchCV, GridSearchCV
# specify parameters and distributions to sample from
param_dist = {"max_depth": [3, None],
"max_features": randint(1,11),
"min_samples_split": randint(2, 11),
"bootstrap": [True, False],
"criterion": ["gini", "entropy"]}
param_dist
{'max_depth': [3, None],
'max_features': <scipy.stats._distn_infrastructure.rv_frozen at 0xf179248>,
'min_samples_split': <scipy.stats._distn_infrastructure.rv_frozen at 0xf1793c8>,
'bootstrap': [True, False],
'criterion': ['gini', 'entropy']}
rf = RandomForestClassifier(n_estimators=20)
n_iter_search = 20
random_search = RandomizedSearchCV(rf, param_distributions=param_dist,
n_iter=n_iter_search, cv=5)
start = time()
random_search.fit(X, Y)
print("RandomizedSearchCV took %.2f seconds for %d candidates"
" parameter settings." % ((time() - start), n_iter_search))
RandomizedSearchCV took 4.52 seconds for 20 candidates parameter settings.
d:\Anaconda3\lib\site-packages\sklearn\model_selection\_search.py:814: DeprecationWarning: The default of the `iid` parameter will change from True to False in version 0.22 and will be removed in 0.24. This will change numeric results when test-set sizes are unequal.
DeprecationWarning)
random_search.best_score_
0.9365609348914858
param_grid = {"max_depth": [3, None],
"max_features": [1, 3, 10],
"min_samples_split": [2, 3, 10],
"bootstrap": [True, False],
"criterion": ["gini", "entropy"]}
# run grid search
grid_search = GridSearchCV(rf, param_grid=param_grid, cv=5)
start = time()
grid_search.fit(X, Y)
print("GridSearchCV took %.2f seconds for %d candidate parameter settings."
% (time() - start, len(grid_search.cv_results_['params'])))
GridSearchCV took 15.34 seconds for 72 candidate parameter settings.
d:\Anaconda3\lib\site-packages\sklearn\model_selection\_search.py:814: DeprecationWarning: The default of the `iid` parameter will change from True to False in version 0.22 and will be removed in 0.24. This will change numeric results when test-set sizes are unequal.
DeprecationWarning)
grid_search.best_score_
0.9354479688369505
- GridSearch & RandomizedSearch can fine tune hyper-parameters of transformers as well when part of pipeline
- GridSearch和RandomizedSearch可以微调变压器的超参数,当管道的一部分时,也可以微调变压器的超参数。
Model Evaluation模型评估
- Three different ways to evaluate quality of model prediction
- score method of estimators, a default method is configured .i.e r2_score for regression, accuracy for classification
- Model evalutaion tools like cross_validate or cross_val_score also returns accuracy
- Metrices module is rich with various prediction error calculation techniques
- 评价模型预测质量的三种不同方法
- 估计器的得分方法,默认配置了一种方法,即r2_score用于回归,准确度用于分类。
- 模型评估工具如cross_validate或cross_val_score等模型评估工具也会返回精度。
- Metrices模块具有丰富的各种预测误差计算技术。
trainX, testX, trainY, testY = train_test_split(X,Y)
rf.fit(trainX, trainY)
RandomForestClassifier(bootstrap=True, class_weight=None, criterion='gini',
max_depth=None, max_features='auto', max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, n_estimators=20,
n_jobs=None, oob_score=False, random_state=None,
verbose=0, warm_start=False)
- Technique 1 - Using score function
- 技巧一----使用评分函数
rf.score(testX,testY)
0.9577777777777777
- Technique 2 - Using cross_val_score as discussed above
- 技巧2 - 使用上文讨论过的cross_val_score
cross_val_score(rf,X,Y,cv=5)
array([0.92307692, 0.90055249, 0.93871866, 0.94677871, 0.88169014])
Cancer prediction sample for understanding metrices癌症预测样本了解度量衡的癌症预测样本
from sklearn.datasets import load_breast_cancer
dt = DecisionTreeClassifier()
cancer_data = load_breast_cancer()
trainX, testX, trainY, testY = train_test_split(cancer_data.data, cancer_data.target)
dt.fit(trainX,trainY)
DecisionTreeClassifier(class_weight=None, criterion='gini', max_depth=None,
max_features=None, max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, presort=False,
random_state=None, splitter='best')
pred = dt.predict(testX)
Technique 3 - Using metrices使用衡量指标
Classfication metrices分类衡量指标
- Accuracy Score - Correct classification vs ( Correct classification + Incorrect Classification )
- 准确率得分 - 正确分类与(正确分类+不正确分类)的比较
from sklearn import metrics
metrics.accuracy_score(y_pred=pred, y_true=testY)
0.9300699300699301
- Confusion Matrix - Shows details of classification inclusing TP,FP,TN,FN
- True Positive (TP), Actual class is 1 & prediction is also 1
- True Negative (TN), Actual class is 0 & prediction is also 0
- False Positive (FP), Acutal class is 0 & prediction is 1
- False Negative (FN), Actual class is 1 & prediction is 0
confusion_result=metrics.confusion_matrix(y_pred=pred, y_true=testY, labels=[0,1])
confusion_result
array([[60, 6],
[ 4, 73]], dtype=int64)
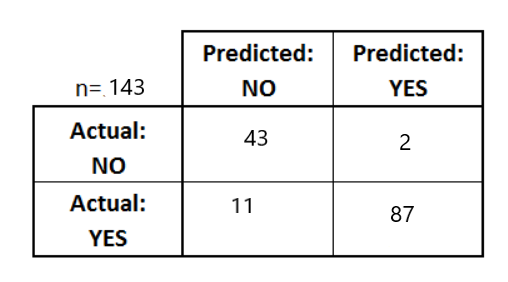
tp=confusion_result[1][1]
tn=confusion_result[0][0]
fp=confusion_result[0][1]
fn=confusion_result[1][0]
- Precision Score精确度得分
- Ability of a classifier not to label positive if the sample is negative
- Claculated as TP/(TP+FP)
- We don’t want a non-spam mail to be marked as spam
- 如果样本为负值,分类器不标记正值的能力
- 按TP/(TP+FP)的形式计算
- 我们不希望非垃圾邮件被标记为垃圾邮件。
precision_result=tp/(tp+fp)
precision_result
0.9240506329113924
metrics.precision_score(y_pred=pred, y_true=testY)
0.9240506329113924
- Recall Score召回率
- Ability of classifier to find all positive samples
- It’s ok to predict patient tumor to be cancer so that it undergoes more test
- But it is not ok to miss a cancer patient without further analysis
- 找到所有阳性样本的能力
- 預測病人的腫瘤是癌症是可以的,所以要多做一些測試
- 但如果没有进一步的分析,错过了癌症患者也是不行的。
metrics.recall_score(y_pred=pred, y_true=testY)
0.948051948051948
- F1 score
- Weighted average of precision & recall
metrics.f1_score(y_pred=pred, y_true=testY)
0.9358974358974359
- ROC & AUC
House Price Prediction - Understanding matrices
from sklearn.datasets import california_housing
house_data = california_housing.fetch_california_housing()
from sklearn.linear_model import LinearRegression
lr = LinearRegression()
lr.fit(house_data.data, house_data.target)
LinearRegression(copy_X=True, fit_intercept=True, n_jobs=None, normalize=False)
pred = lr.predict(house_data.data)
Matrices for Regression
- mean squared error
- Sum of squares of difference between expected value & actual value
metrics.mean_squared_error(y_pred=pred, y_true=house_data.target)
0.5243209861846071
- mean absolute error
- Sum of abs of difference between expected value & actual value
metrics.mean_absolute_error(y_pred=pred, y_true=house_data.target)
0.5311643817546461
-
r2 score
- Returns accuracy of model in the scale of 0 & 1
- It measures goodness of fit for regression models
- Calculated as = (variance explained by the model)/(Total variance)
- High r2 means target is close to prediction
metrics.r2_score(y_pred=pred, y_true=house_data.target)
0.6062326851998051
Metrices for Clustering 用于聚类的衡量指标
- Two forms of evaluation
- supervised, which uses a ground truth class values for each sample.
- completeness_score
- homogeneity_score
- unsupervised, which measures the quality of model itself
- silhoutte_score
- calinski_harabaz_score
- 两种评价形式
- 监督的,它为每个样本使用了一个地面真值类的值。
- 完整度_score
- 同质性分数
- 无监督,衡量模型本身的质量
- silhoutte_score
- calinski_harabaz_score
completeness_score
- A clustering result satisfies completeness if all the data points that are members of a given class are elements of the same cluster.
- Accuracy is 1.0 if data belonging to same class belongs to same cluster, even if multiple classes belongs to same cluster
from sklearn.metrics.cluster import completeness_score
completeness_score( labels_true=[10,10,11,11],labels_pred=[1,1,0,0])
1.0
- The acuracy is 1.0 because all the data belonging to same class belongs to same cluster
completeness_score( labels_true=[11,22,22,11],labels_pred=[1,0,1,1])
0.3836885465963443
- The accuracy is .3 because class 1 - [11,22,11], class 2 - [22]
print(completeness_score([10, 10, 11, 11], [0, 0, 0, 0]))
1.0
homogeneity_score
- A clustering result satisfies homogeneity if all of its clusters contain only data points which are members of a single class.
from sklearn.metrics.cluster import homogeneity_score
homogeneity_score([0, 0, 1, 1], [1, 1, 0, 0])
1.0
homogeneity_score([0, 0, 1, 1], [0, 1, 2, 3])
0.9999999999999999
homogeneity_score([0, 0, 0, 0], [1, 1, 0, 0])
1.0
- Same class data is broken into two clusters
silhoutte_score
- The Silhouette Coefficient is calculated using the mean intra-cluster distance (a) and the mean nearest-cluster distance (b) for each sample.
- The Silhouette Coefficient for a sample is (b - a) / max(a, b). To clarify, b is the distance between a sample and the nearest cluster that the sample is not a part of.
Selecting the number of clusters with silhouette analysis on KMeans clustering
from sklearn.datasets import make_blobs
X, Y = make_blobs(n_samples=500,
n_features=2,
centers=4,
cluster_std=1,
center_box=(-10.0, 10.0),
shuffle=True,
random_state=1)
plt.scatter(X[:,0],X[:,1],s=10)

range_n_clusters = [2, 3, 4, 5, 6]
from sklearn.cluster import KMeans
from sklearn.metrics import silhouette_score
for n_cluster in range_n_clusters:
kmeans = KMeans(n_clusters=n_cluster)
kmeans.fit(X)
labels = kmeans.predict(X)
print (n_cluster, silhouette_score(X,labels))
2 0.7049787496083262
3 0.5882004012129721
4 0.6505186632729437
5 0.5746932321727457
6 0.49417400746431644
- The best number of clusters is 2
calinski_harabaz_score
- The score is defined as ratio between the within-cluster dispersion and the between-cluster dispersion.
from sklearn.metrics import calinski_harabaz_score
for n_cluster in range_n_clusters:
kmeans = KMeans(n_clusters=n_cluster)
kmeans.fit(X)
labels = kmeans.predict(X)
print (n_cluster, calinski_harabaz_score(X,labels))
2 1604.112286409658
3 1809.991966958033
4 2704.4858735121097
5 2281.91411035916
6 2040.6320809618921
Model Persistance
- Model training is an expensive process
- It is desireable to save the model for future reuse
- using pickle & joblib this can be achieved
- 模型培训是一个昂贵的过程
- 希望将模型保存下来,以便将来再利用。
- 使用pickle和joblib可以实现这个功能
import pickle
s = pickle.dumps(dt)
pickle.loads(s)
DecisionTreeClassifier(class_weight=None, criterion='gini', max_depth=None,
max_features=None, max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, presort=False,
random_state=None, splitter='best')
type(s)
bytes
- joblib is better extension of pickle
- Doesn’t convert into string
from sklearn.externals import joblib
joblib.dump(dt, 'dt.joblib')
['dt.joblib']
- Loading the file back into model
dt = joblib.load('dt.joblib')
dt
DecisionTreeClassifier(class_weight=None, criterion='gini', max_depth=None,
max_features=None, max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, presort=False,
random_state=None, splitter='best')
Validation Curves验证曲线
- To validate a model, we need a scoring function.
- Create a grid of possible hyper-prameter configuration.
- Select the hyper-parameter which gives the best score
- 要验证一个模型,我们需要一个评分函数。
- 创建一个可能的超参数配置的网格。
- 选择一个能给出最佳得分的超参数。
from sklearn.model_selection import validation_curve
param_range = np.arange(1, 50, 2)
train_scores, test_scores = validation_curve(RandomForestClassifier(),
digits.data,
digits.target,
param_name="n_estimators",
param_range=param_range,
cv=3,
scoring="accuracy",
n_jobs=-1)
train_mean = np.mean(train_scores, axis=1)
train_std = np.std(train_scores, axis=1)
test_mean = np.mean(test_scores, axis=1)
test_std = np.std(test_scores, axis=1)
plt.plot(param_range, train_mean, label="Training score", color="black")
plt.plot(param_range, test_mean, label="Cross-validation score", color="dimgrey")
plt.title("Validation Curve With Random Forest")
plt.xlabel("Number Of Trees")
plt.ylabel("Accuracy Score")
plt.tight_layout()
plt.legend(loc="best")
plt.show()

6. Learning Curves
- Learning curves shows variation in training & validation score on increasing the number of samples
from sklearn.model_selection import learning_curve
