Spark 分布式实现线性判别分析(二分类问题)
线性判别分析(LDA)
线性判别分析(linear discriminant analysis)是一种经典的线性学习方法,在二分类问题上最早由Fisher在1936年提出,亦称Fisher线性判别。其中心思想是:将样本集投影到一条直线上,使投影到直线上的同类样本之间差异尽可能的小;使不同类样本间的差异尽可能的大。线性判别分析是一种有监督的分类学习方法;LDA也可以用于数据降维处理。
算法步骤
1 计算各个类的样本均值;
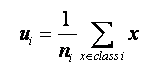
2.计算总体样本均值;
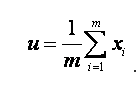
3.计算组内协方差矩阵与组间协方差矩阵
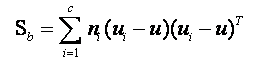
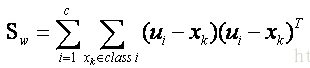
4.求线性判别函数
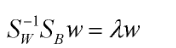
求出Sw^(-1) Sb 的 最大特征向量即为线性判别函数的系数
相关证明及更好的教程请参见
数据展示与说明
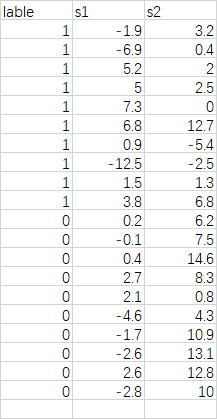
本数据来源于《多元统计分析及R语言建模》书中的案例数据,数据描述如下:
今天和昨天的湿温差s1以及气温差s2是预报明天下雨或不下雨的两个重要因子。0表示不下雨,1表示下雨。本案例将针对该数据建立fisher线性判别模型对数据进行判别分析。
实现代码
我们先自定义一个计算样本均值向量的自定义聚合函数
import org.apache.spark.sql.expressions.{
MutableAggregationBuffer,
UserDefinedAggregateFunction
}
import org.apache.spark.sql.types._
/**
* Created by WZZC on 2019/2/27
**/
class meanVector(len: Long) extends UserDefinedAggregateFunction {
override def inputSchema: StructType =
new StructType()
.add("features", DataTypes.createArrayType(DoubleType))
override def bufferSchema: StructType =
new StructType()
.add("features", DataTypes.createArrayType(DoubleType))
.add("count", LongType)
override def dataType: DataType = DataTypes.createArrayType(DoubleType)
override def deterministic: Boolean = true
override def initialize(buffer: MutableAggregationBuffer): Unit = {
buffer.update(0, new Array[Double](len.toInt).toSeq)
buffer.update(1, 0L)
}
override def update(buffer: MutableAggregationBuffer, input: Row): Unit = {
val inputVec = input.getSeq[Double](0)
val bufferVec = buffer.getSeq[Double](0)
val outputVec: Seq[Double] = if (bufferVec.length == 1) {
inputVec
} else {
inputVec.zip(bufferVec).map(x => x._1 + x._2)
}
buffer.update(0, outputVec)
buffer.update(1, buffer.getLong(1) + 1)
}
override def merge(buffer1: MutableAggregationBuffer, buffer2: Row): Unit = {
val vec1 = buffer1.getSeq[Double](0)
val vec2 = buffer2.getSeq[Double](0)
val newVec = vec1.zip(vec2).map(x => x._1 + x._2)
buffer1.update(0, newVec)
buffer1.update(1, buffer1.getLong(1) + buffer2.getLong(1))
}
override def evaluate(buffer: Row): Any = {
val length = buffer.getLong(1)
buffer.getSeq[Double](0).map(_ / length)
}
}
def main(args: Array[String]): Unit = {
val spark = SparkSession
.builder()
.appName(s"${this.getClass.getSimpleName}")
.master("local[*]")
.getOrCreate()
val ldaData = spark.read
.option("inferSchema", true)
.csv("F:\\DataSource\\lda.csv")
.toDF("lable", "s1", "s2")
import spark.implicits._
val ldaschema = ldaData.schema
val fts = ldaschema.filterNot(_.name == "lable").map(_.name).toArray
val amountVectorAssembler: VectorAssembler = new VectorAssembler()
.setInputCols(fts)
.setOutputCol("features")
val vec2Array = udf((vec: DenseVector) => vec.toArray)
val ldaFeatrus = amountVectorAssembler
.transform(ldaData)
.select($"lable", vec2Array($"features") as "features")
val meanVector = spark.udf.register("meanVector", new meanVector(fts.length))
val uiGroup = ldaFeatrus
.groupBy($"lable")
.agg(meanVector($"features") as "ui", count($"lable") as "len")
// 类别、协方差矩阵、均值向量 、长度
val covMatrix = ldaFeatrus
.join(uiGroup, "lable")
.rdd
.map(row => {
val lable = row.getAs[Int]("lable")
val len = row.getAs[Long]("len")
val u = densevec(row.getAs[Seq[Double]]("ui").toArray)
val x = densevec(row.getAs[Seq[Double]]("features").toArray)
val denseMatrix = (x - u).toDenseMatrix
lable -> (denseMatrix, u, len)
})
.reduceByKey((d1, d2) => {
(DenseMatrix.vertcat(d1._1, d2._1), d1._2, d1._3)
})
.map(tp => {
val len = tp._2._3 - 1
val t: DenseMatrix[Double] = (tp._2._1.t * tp._2._1).map(x => x / len)
(tp._1, t, tp._2._2.toDenseMatrix, len + 1)
})
// 总体均值向量
val uaVec = covMatrix.map(x => (x._3.toDenseVector, x._4))
.reduce((u1, u2) => {
val ua = u1._1.map(_ * u1._2) + u2._1.map(_ * u2._2)
ua.map(_ / (u1._2 + u2._2)) -> 1
})
._1.toDenseMatrix
val (a, sw, sb, b) = covMatrix.reduce((c1, c2) => {
val sw = c1._2 + c2._2 //
val d = (c1._3.toDenseVector - c2._3.toDenseVector).toDenseMatrix
val sb = d.t * d //
(1, sw, sb, 1L)
})
// 计算特征值与特征向量
val eigs = eig(inv(sw) * sb)
// 最大特征值
val maxEigenvalues = bMax(eigs.eigenvalues)
val maxEigenvaluesIndex = eigs.eigenvalues.data.indexOf(maxEigenvalues)
// 提取最大特征向量
val maxEigenvectors = eigs.eigenvectors(::, maxEigenvaluesIndex)
// 计算分隔点
val separationPoint = (uaVec * maxEigenvectors).data.head
val xbars = covMatrix.map(x => {
val xbar = (maxEigenvectors.toDenseMatrix * x._3.t).data.head
val bool = (xbar > separationPoint).toString
if (xbar == separationPoint) "n" -> -1d else bool -> x._1.toDouble
}).collect().toMap
// 定义判别函数
val predictudf = udf((seq: Seq[Double]) => {
val y = (densevec(seq.toArray).toDenseMatrix * maxEigenvectors).data.head
val bool = (y > separationPoint).toString
if (y - separationPoint == 0) {
-1d
} else {
xbars.getOrElse(bool, -1d)
}
})
val predictions = ldaFeatrus.withColumn("newL", predictudf($"features"))
predictions.show()
spark.stop()
}
判别结果展示
+-----+------------+----+
|lable| features|newL|
+-----+------------+----+
| 1| [-1.9,3.2]| 1.0|
| 1| [-6.9,0.4]| 1.0|
| 1| [5.2,2.0]| 1.0|
| 1| [5.0,2.5]| 1.0|
| 1| [7.3,0.0]| 1.0|
| 1| [6.8,12.7]| 0.0|
| 1| [0.9,-5.4]| 1.0|
| 1|[-12.5,-2.5]| 1.0|
| 1| [1.5,1.3]| 1.0|
| 1| [3.8,6.8]| 1.0|
| 0| [0.2,6.2]| 0.0|
| 0| [-0.1,7.5]| 0.0|
| 0| [0.4,14.6]| 0.0|
| 0| [2.7,8.3]| 0.0|
| 0| [2.1,0.8]| 1.0|
| 0| [-4.6,4.3]| 0.0|
| 0| [-1.7,10.9]| 0.0|
| 0| [-2.6,13.1]| 0.0|
| 0| [2.6,12.8]| 0.0|
| 0| [-2.8,10.0]| 0.0|
+-----+------------+----+
对于模型好坏的判别,我们可以通过混淆矩阵进行分析,Spark中也提供了相关指标的计算工具,在此就以准确率与召回率为例。
// 正确率
val evaluator1 = new MulticlassClassificationEvaluator()
.setLabelCol("lable")
.setPredictionCol("newL")
.setMetricName("accuracy")
val accuracy = evaluator1.evaluate(predictions)
println("正确率 =" + accuracy)
// 召回率
val evaluator2 = new MulticlassClassificationEvaluator()
.setLabelCol("lable")
.setPredictionCol("newL")
.setMetricName("accuracy")
val Recall = evaluator2.evaluate(predictions)
println("召回率 = " + Recall)
正确率 =0.9
召回率 = 0.9
参考资料
https://blog.csdn.net/qq_20406597/article/details/80166589
https://blog.csdn.net/jnulzl/article/details/49894041
《多元统计分析及R语言建模》 – 王斌会