Programming Exercise 7.2:Principal Component Analysis(PCA)
Python版本3.6
编译环境:anaconda Jupyter Notebook
链接:实验数据和实验指导书
提取码:i7co
本章课程笔记部分见:14.降维
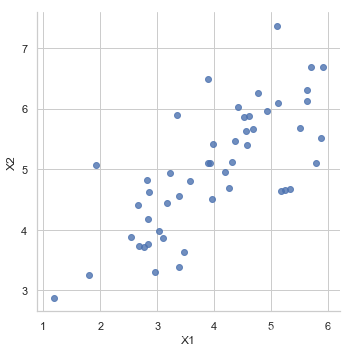
PCA是在数据集中找到“主成分”或最大方差方向的线性变换。 它可以用于降维。 在本练习中,我们首先负责实现PCA并将其应用于一个简单的二维数据集,以了解它是如何工作的。 我们从加载和可视化数据集开始。
%matplotlib inline
#IPython的内置magic函数,可以省掉plt.show(),在其他IDE中是不会支持的
import numpy as np
import pandas as pd
import matplotlib as mpl
import matplotlib.pyplot as plt
import seaborn as sns
import scipy.io as sio
sns.set(style="whitegrid",color_codes=True)
4-二维的PCA
mat = sio.loadmat('./data/ex7data1.mat')
X = mat['X']
def get_X(df):
"""
use concat to add intersect feature to avoid side effect
not efficient for big dataset though
"""
ones = pd.DataFrame({'ones': np.ones(len(df))})
data = pd.concat([ones, df], axis=1) # column concat
return data.iloc[:, :-1].as_matrix() # this return ndarray, not matrix
def get_y(df):
'''assume the last column is the target'''
return np.array(df.iloc[:, -1])
def normalize_feature(df):
"""Applies function along input axis(default 0) of DataFrame."""
return df.apply(lambda column: (column - column.mean()) / column.std())
sns.lmplot('X1', 'X2',
data=pd.DataFrame(X, columns=['X1', 'X2']),
fit_reg=False)
<seaborn.axisgrid.FacetGrid at 0x8bf2e6c9e8>
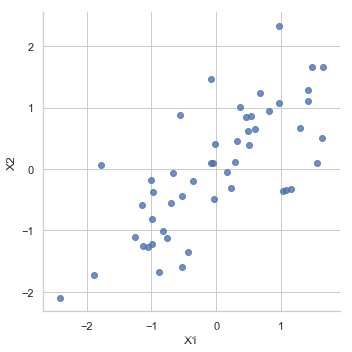
normalize data
# support functions ---------------------------------------
def plot_n_image(X, n):
""" plot first n images
n has to be a square number
"""
pic_size = int(np.sqrt(X.shape[1]))
grid_size = int(np.sqrt(n))
first_n_images = X[:n, :]
fig, ax_array = plt.subplots(nrows=grid_size, ncols=grid_size,
sharey=True, sharex=True, figsize=(8, 8))
for r in range(grid_size):
for c in range(grid_size):
ax_array[r, c].imshow(first_n_images[grid_size * r + c].reshape((pic_size, pic_size)))
plt.xticks(np.array([]))
plt.yticks(np.array([]))
# PCA functions ---------------------------------------
def covariance_matrix(X):
"""
Args:
X (ndarray) (m, n)
Return:
cov_mat (ndarray) (n, n):
covariance matrix of X
"""
m = X.shape[0]
return (X.T @ X) / m
def normalize(X):
"""
for each column, X-mean / std
"""
X_copy = X.copy()
m, n = X_copy.shape
for col in range(n):
X_copy[:, col] = (X_copy[:, col] - X_copy[:, col].mean()) / X_copy[:, col].std()
return X_copy
def pca(X):
"""
http://docs.scipy.org/doc/numpy/reference/generated/numpy.linalg.svd.html
Args:
X ndarray(m, n)
Returns:
U ndarray(n, n): principle components
"""
# 1. normalize data
X_norm = normalize(X)
# 2. calculate covariance matrix
Sigma = covariance_matrix(X_norm) # (n, n)
# 3. do singular value decomposition
# remeber, we feed cov matrix in SVD, since the cov matrix is symmetry
# left sigular vector and right singular vector is the same, which means
# U is V, so we could use either one to do dim reduction
U, S, V = np.linalg.svd(Sigma) # U: principle components (n, n)
return U, S, V
def project_data(X, U, k):
"""
Args:
U (ndarray) (n, n)
Return:
projected X (n dim) at k dim
"""
m, n = X.shape
if k > n:
raise ValueError('k should be lower dimension of n')
return X @ U[:, :k]
def recover_data(Z, U):
m, n = Z.shape
if n >= U.shape[0]:
raise ValueError('Z dimension is >= U, you should recover from lower dimension to higher')
return Z @ U[:, :n].T
X_norm = normalize(X)
sns.lmplot('X1', 'X2',
data=pd.DataFrame(X_norm, columns=['X1', 'X2']),
fit_reg=False)
<seaborn.axisgrid.FacetGrid at 0x8be9d90c18>
covariance matrix
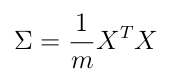
Sigma =covariance_matrix(X_norm) # capital greek Sigma
Sigma # (n, n)
array([[1. , 0.73553038],
[0.73553038, 1. ]])
PCA
U, S, V = pca(X_norm)
project data to lower dimension
Z = project_data(X_norm, U, 1)
Z[:10]
array([[ 1.49631261],
[-0.92218067],
[ 1.22439232],
[ 1.64386173],
[ 1.2732206 ],
[-0.97681976],
[ 1.26881187],
[-2.34148278],
[-0.02999141],
[-0.78171789]])
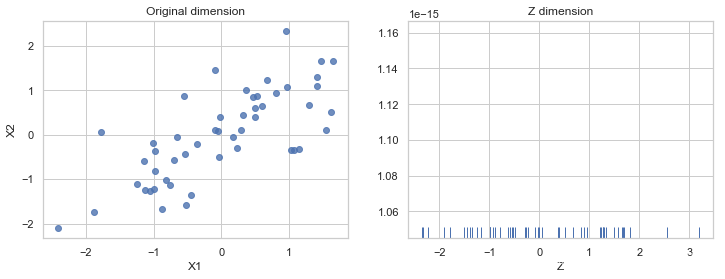
fig, (ax1, ax2) = plt.subplots(ncols=2, figsize=(12, 4))
sns.regplot('X1', 'X2',
data=pd.DataFrame(X_norm, columns=['X1', 'X2']),
fit_reg=False,
ax=ax1)
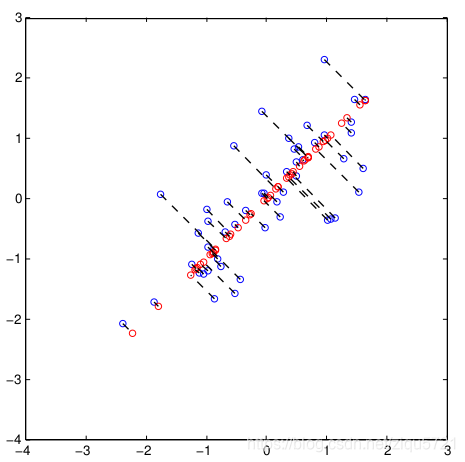
ax1.set_title('Original dimension')
sns.rugplot(Z, ax=ax2)
ax2.set_xlabel('Z')
ax2.set_title('Z dimension')
Text(0.5, 1.0, 'Z dimension')
recover data to original dimension
X_recover = recover_data(Z, U)
fig, (ax1, ax2, ax3) = plt.subplots(ncols=3, figsize=(12, 4))
sns.rugplot(Z, ax=ax1)
ax1.set_title('Z dimension')
ax1.set_xlabel('Z')
sns.regplot('X1', 'X2',
data=pd.DataFrame(X_recover, columns=['X1', 'X2']),
fit_reg=False,
ax=ax2)
ax2.set_title("2D projection from Z")
sns.regplot('X1', 'X2',
data=pd.DataFrame(X_norm, columns=['X1', 'X2']),
fit_reg=False,
ax=ax3)
ax3.set_title('Original dimension')
Text(0.5, 1.0, 'Original dimension')

the projection from (X1, X2) to Z could be visualized like this
5-PCA用于面部数据
此练习中的最后一个任务是将PCA应用于脸部图像。 通过使用相同的降维技术,我们可以使用比原始图像少得多的数据来捕获图像的“本质”。
mat = sio.loadmat('./data/ex7faces.mat')
mat.keys()
dict_keys(['__header__', '__version__', '__globals__', 'X'])
X = np.array([x.reshape((32, 32)).T.reshape(1024) for x in mat.get('X')])
X.shape
(5000, 1024)
# support functions ---------------------------------------
def plot_n_image(X, n):
""" plot first n images
n has to be a square number
"""
pic_size = int(np.sqrt(X.shape[1]))
grid_size = int(np.sqrt(n))
first_n_images = X[:n, :]
fig, ax_array = plt.subplots(nrows=grid_size, ncols=grid_size,
sharey=True, sharex=True, figsize=(8, 8))
for r in range(grid_size):
for c in range(grid_size):
ax_array[r, c].imshow(first_n_images[grid_size * r + c].reshape((pic_size, pic_size)))
plt.xticks(np.array([]))
plt.yticks(np.array([]))
# PCA functions ---------------------------------------
def covariance_matrix(X):
"""
Args:
X (ndarray) (m, n)
Return:
cov_mat (ndarray) (n, n):
covariance matrix of X
"""
m = X.shape[0]
return (X.T @ X) / m
def normalize(X):
"""
for each column, X-mean / std
"""
X_copy = X.copy()
m, n = X_copy.shape
for col in range(n):
X_copy[:, col] = (X_copy[:, col] - X_copy[:, col].mean()) / X_copy[:, col].std()
return X_copy
def pca(X):
"""
http://docs.scipy.org/doc/numpy/reference/generated/numpy.linalg.svd.html
Args:
X ndarray(m, n)
Returns:
U ndarray(n, n): principle components
"""
# 1. normalize data
X_norm = normalize(X)
# 2. calculate covariance matrix
Sigma = covariance_matrix(X_norm) # (n, n)
# 3. do singular value decomposition
# remeber, we feed cov matrix in SVD, since the cov matrix is symmetry
# left sigular vector and right singular vector is the same, which means
# U is V, so we could use either one to do dim reduction
U, S, V = np.linalg.svd(Sigma) # U: principle components (n, n)
return U, S, V
def project_data(X, U, k):
"""
Args:
U (ndarray) (n, n)
Return:
projected X (n dim) at k dim
"""
m, n = X.shape
if k > n:
raise ValueError('k should be lower dimension of n')
return X @ U[:, :k]
def recover_data(Z, U):
m, n = Z.shape
if n >= U.shape[0]:
raise ValueError('Z dimension is >= U, you should recover from lower dimension to higher')
return Z @ U[:, :n].T
plot_n_image(X, n=64)

run PCA, find principle components
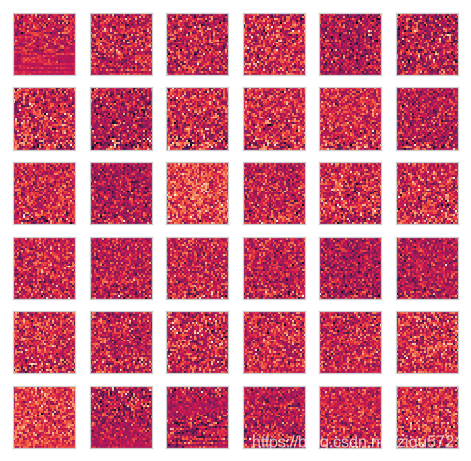
U, _, _ = pca(X)
U.shape
(1024, 1024)
plot_n_image(U, n=36)
reduce dimension to k=100
Z = project_data(X, U, k=100)
plot_n_image(Z, n=64)

recover from k=100
X_recover = recover_data(Z, U)
plot_n_image(X_recover, n=64)

sklearn PCA
from sklearn.decomposition import PCA
sk_pca = PCA(n_components=100)
Z = sk_pca.fit_transform(X)
Z.shape
(5000, 100)
plot_n_image(Z, 64)

X_recover = sk_pca.inverse_transform(Z)
X_recover.shape
(5000, 1024)
plot_n_image(X_recover, n=64)
