目录
1.单个变量异常值检测
如果某个值离平均值有多个标准偏差,并且远离近似标准正态分布的值,则我们更倾向于将将其视为异常值。
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import statsmodels.api as sm
import scipy.stats as scistat
data=pd.read_csv("F:\\python数据清洗\\04第四章\\covidtotals.csv")data.columnsIndex(['iso_code', 'lastdate', 'location', 'total_cases', 'total_deaths',
'total_cases_pm', 'total_deaths_pm', 'population', 'pop_density',
'median_age', 'gdp_per_capita', 'hosp_beds'],
dtype='object')
data.set_index('iso_code',inplace=True)
totvars=['location', 'total_cases', 'total_deaths', 'total_cases_pm', 'total_deaths_pm']
demovars=['population', 'pop_density', 'median_age', 'gdp_per_capita', 'hosp_beds']1.1 描述性统计
data_b=data.loc[:,totvars]
data_b.describe()| total_cases | total_deaths | total_cases_pm | total_deaths_pm | |
|---|---|---|---|---|
| count | 2.100000e+02 | 210.000000 | 210.000000 | 210.000000 |
| mean | 2.921614e+04 | 1770.714286 | 1355.357943 | 55.659129 |
| std | 1.363978e+05 | 8705.565857 | 2625.277497 | 144.785816 |
| min | 0.000000e+00 | 0.000000 | 0.000000 | 0.000000 |
| 25% | 1.757500e+02 | 4.000000 | 92.541500 | 0.884750 |
| 50% | 1.242500e+03 | 25.500000 | 280.928500 | 6.154000 |
| 75% | 1.011700e+04 | 241.250000 | 1801.394750 | 31.777250 |
| max | 1.790191e+06 | 104383.000000 | 19771.348000 | 1237.551000 |
1.2 显示百分位数数据
data_b.quantile(np.arange(0.0,1.1,0.1))| total_cases | total_deaths | total_cases_pm | total_deaths_pm | |
|---|---|---|---|---|
| 0.0 | 0.0 | 0.0 | 0.0000 | 0.0000 |
| 0.1 | 22.9 | 0.0 | 17.9986 | 0.0000 |
| 0.2 | 105.2 | 2.0 | 56.2910 | 0.3752 |
| 0.3 | 302.0 | 6.7 | 115.4341 | 1.7183 |
| 0.4 | 762.0 | 12.0 | 213.9734 | 3.9566 |
| 0.5 | 1242.5 | 25.5 | 280.9285 | 6.1540 |
| 0.6 | 2514.6 | 54.6 | 543.9562 | 12.2452 |
| 0.7 | 6959.8 | 137.2 | 1071.2442 | 25.9459 |
| 0.8 | 16847.2 | 323.2 | 2206.2982 | 49.9658 |
| 0.9 | 46513.1 | 1616.9 | 3765.1363 | 138.9045 |
| 1.0 | 1790191.0 | 104383.0 | 19771.3480 | 1237.5510 |
1.3 偏度(skewness)&峰度(kurtosis)
偏度和峰度分别描述了分布的对称性和分布的尾部有多肥。如果变量以正态分布,则这两个指标都大大高于预期。
# 偏度skewness
data_b.skew()
----------------------------
total_cases 10.804275
total_deaths 8.929816
total_cases_pm 4.396091
total_deaths_pm 4.674417
dtype: float64
# 峰度kurtosis
data_b.kurtosis()
----------------------------
total_cases 134.979577
total_deaths 95.737841
total_cases_pm 25.242790
total_deaths_pm 27.238232
dtype: float641.4 测试数据的正态分布性
使用scipy库中的Shapiro-Wilk测试,输出测试的p值。正态分布的null假设可以在低于0.05的任何p值和95%的置信水平上被拒绝。
def testnorm(var,df):
stat,p=scistat.shapiro(df[var])
return p
testnorm("total_cases",data_b) #3.753789128593843e-29
testnorm("total_deaths",data_b) #4.3427896631016077e-29
testnorm("total_cases_pm",data_b) #1.3972683006509067e-23
testnorm("total_deaths_pm",data_b) #1.361060423265974e-25结果表明变量的分布在高显著水平下不是正态的。
# 总病例数的分位数图(qq图),直线显示的是正态分布应有的外观
sm.qqplot(data_b[["total_cases"]].sort_values(['total_cases']),line='s')
plt.title('QQ Plot of Total Cases')
plt.show()
# 每百万人口的总病例数的分位数图(qq图)
sm.qqplot(data_b[["total_cases_pm"]].sort_values(['total_cases_pm']),line='s')
plt.title('QQ Plot of Total Cases Per Million')
plt.show() 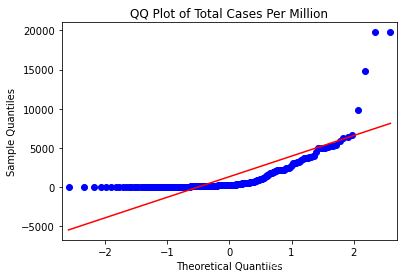
qqplot的结果也表明,total_cases(总病例数)和total_cases_pm(每百万人口的总病例数)的分布都与正态分布有显著差异(红色直线显示的是正态分布应有的外观)
1.5 显示离群值范围
定义变量离群值的一种方法是按四分位数的距离计算。第三个四分位数(0.75)被称为上四分位数,第一个四分位数(0.25)被称为下四分位数,上四分位数和下四分位数之间的距离称为四分位距,也就是0.25~0.75分位数的范围。如果某个值离均值大于1.5倍,则认为该值是离群值。
def getoutliers():
dfout=pd.DataFrame(columns=data.columns,data=None)
for col in data_b.columns[1:]:
thirdq,firstq=data_b[col].quantile(0.75),data_b[col].quantile(0.25) #计算四分位数
range_q=1.5*(thirdq-firstq) #四分位距
high,low=range_q+thirdq,firstq-range_q #离群阈值
df=data.loc[(data[col]>high) | (data[col]<low)] #筛选出离群点
df=df.assign(varname=col,threshlow=low,threshhigh=high) #添加新列来显示所检查的变量(varname)和离群范围
dfout=pd.concat([dfout,df]) #合并表格
return dfout
outliers=getoutliers()
outliers| lastdate | location | total_cases | total_deaths | total_cases_pm | total_deaths_pm | population | pop_density | median_age | gdp_per_capita | hosp_beds | varname | threshlow | threshhigh | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BGD | 2020-06-01 | Bangladesh | 47153 | 650 | 286.315 | 3.947 | 164689383.0 | 1265.036 | 27.5 | 3523.984 | 0.80 | total_cases | -14736.125 | 25028.875 |
| BLR | 2020-06-01 | Belarus | 42556 | 235 | 4503.604 | 24.870 | 9449321.0 | 46.858 | 40.3 | 17167.967 | 11.00 | total_cases | -14736.125 | 25028.875 |
| BEL | 2020-06-01 | Belgium | 58381 | 9467 | 5037.354 | 816.852 | 11589616.0 | 375.564 | 41.8 | 42658.576 | 5.64 | total_cases | -14736.125 | 25028.875 |
| BRA | 2020-06-01 | Brazil | 514849 | 29314 | 2422.142 | 137.910 | 212559409.0 | 25.040 | 33.5 | 14103.452 | 2.20 | total_cases | -14736.125 | 25028.875 |
| CAN | 2020-06-01 | Canada | 90936 | 7295 | 2409.401 | 193.285 | 37742157.0 | 4.037 | 41.4 | 44017.591 | 2.50 | total_cases | -14736.125 | 25028.875 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| ESP | 2020-05-31 | Spain | 239429 | 27127 | 5120.952 | 580.197 | 46754783.0 | 93.105 | 45.5 | 34272.360 | 2.97 | total_deaths_pm | -45.454 | 78.116 |
| SWE | 2020-06-01 | Sweden | 37542 | 4395 | 3717.298 | 435.180 | 10099270.0 | 24.718 | 41.0 | 46949.283 | 2.22 | total_deaths_pm | -45.454 | 78.116 |
| CHE | 2020-06-01 | Switzerland | 30779 | 1656 | 3556.367 | 191.343 | 8654618.0 | 214.243 | 43.1 | 57410.166 | 4.53 | total_deaths_pm | -45.454 | 78.116 |
| GBR | 2020-06-01 | United Kingdom | 274762 | 38489 | 4047.403 | 566.965 | 67886004.0 | 272.898 | 40.8 | 39753.244 | 2.54 | total_deaths_pm | -45.454 | 78.116 |
| USA | 2020-06-01 | United States | 1790191 | 104383 | 5408.389 | 315.354 | 331002647.0 | 35.608 | 38.3 | 54225.446 | 2.77 | total_deaths_pm | -45.454 | 78.116 |
outliers.varname.value_counts()total_deaths 36 total_cases 33 total_deaths_pm 28 total_cases_pm 17 Name: varname, dtype: int64
2. 双变量关系中的异常值检测
当某个值的分布与平均值没有明显偏离时,即使它不是极值,它也可能是异常值。当第二个变量具有某些值时,第一个变量的某些值就可能是异常值。(以下使用的还是之前的数据)
2.1 生成相关性矩阵
data.corr(method="pearson")| total_cases | total_deaths | total_cases_pm | total_deaths_pm | population | pop_density | median_age | gdp_per_capita | hosp_beds | |
|---|---|---|---|---|---|---|---|---|---|
| total_cases | 1.000000 | 0.932079 | 0.182246 | 0.247464 | 0.270030 | -0.028737 | 0.162698 | 0.186835 | 0.027601 |
| total_deaths | 0.932079 | 1.000000 | 0.179812 | 0.394811 | 0.212619 | -0.031645 | 0.205128 | 0.198729 | 0.019990 |
| total_cases_pm | 0.182246 | 0.179812 | 1.000000 | 0.586468 | -0.056009 | 0.110043 | 0.313836 | 0.651200 | 0.081449 |
| total_deaths_pm | 0.247464 | 0.394811 | 0.586468 | 1.000000 | -0.013902 | 0.030281 | 0.389595 | 0.383672 | 0.120488 |
| population | 0.270030 | 0.212619 | -0.056009 | -0.013902 | 1.000000 | -0.023084 | 0.024395 | -0.059555 | -0.038329 |
| pop_density | -0.028737 | -0.031645 | 0.110043 | 0.030281 | -0.023084 | 1.000000 | 0.178878 | 0.315199 | 0.314973 |
| median_age | 0.162698 | 0.205128 | 0.313836 | 0.389595 | 0.024395 | 0.178878 | 1.000000 | 0.648905 | 0.662222 |
| gdp_per_capita | 0.186835 | 0.198729 | 0.651200 | 0.383672 | -0.059555 | 0.315199 | 0.648905 | 1.000000 | 0.296995 |
| hosp_beds | 0.027601 | 0.019990 | 0.081449 | 0.120488 | -0.038329 | 0.314973 | 0.662222 | 0.296995 | 1.000000 |
从上表可看出,总病例数与总死亡数之间的相关性非常高(0.93),而每百万人口总病例数与每百万人口总死亡数之间的相关性则较小(0.59),人均GDP与每百万人口总病例数之间也存在较强的相关关系(0.65)。
2.2 创建不同变量分位数的交叉表
首先创建一个DataFrame,使用qcut创建一个将数据分解为分位数的列,显示总病例数分位数与总死亡数数分位数的交叉表。
data_b['total_cases_q']=pd.qcut(data_b['total_cases'],labels=['very low','low','medium','high','very high'],q=5,precision=0)
data_b['total_deaths_q']=pd.qcut(data_b['total_deaths'],labels=['very low','low','medium','high','very high'],q=5,precision=0)
pd.crosstab(data_b.total_cases_q,data_b.total_deaths_q)| total_deaths_q | very low | low | medium | high | very high |
|---|---|---|---|---|---|
| total_cases_q | |||||
| very low | 34 | 7 | 1 | 0 | 0 |
| low | 12 | 19 | 10 | 1 | 0 |
| medium | 1 | 13 | 15 | 13 | 0 |
| high | 0 | 0 | 12 | 24 | 6 |
| very high | 0 | 0 | 2 | 4 | 36 |
data.loc[(data_b.total_cases_q=="very high")&(data_b.total_deaths_q=="medium")].T| iso_code | QAT | SGP |
|---|---|---|
| lastdate | 2020-06-01 | 2020-06-01 |
| location | Qatar | Singapore |
| total_cases | 56910 | 34884 |
| total_deaths | 38 | 23 |
| total_cases_pm | 19753.146 | 5962.727 |
| total_deaths_pm | 13.19 | 3.931 |
| population | 2881060.0 | 5850343.0 |
| pop_density | 227.322 | 7915.731 |
| median_age | 31.9 | 42.4 |
| gdp_per_capita | 116935.6 | 85535.383 |
| hosp_beds | 1.2 | 2.4 |
从表中可以看出,大多数国家都在对角线上或其附近,但有2个国家(卡塔尔,新加坡)的病例数很高,但死亡人数时中等的。
2.3 绘制两个变量间的散点图
使用seaborn的regplot方法时,除生成散点图外,还可以生成线性回归线。
import seaborn as sns
ax=sns.regplot(x="total_cases",y="total_deaths",data=data)
ax.set(xlabel='Cases',ylabel='Deaths',title='Total Covid Cases and Deaths by Country')
plt.show()
# 检查回归线上方的意外值
data.loc[(data.total_cases<300000) & (data.total_deaths>20000)].T| iso_code | FRA | ITA | ESP | GBR |
|---|---|---|---|---|
| lastdate | 2020-06-01 | 2020-06-01 | 2020-05-31 | 2020-06-01 |
| location | France | Italy | Spain | United Kingdom |
| total_cases | 151753 | 233019 | 239429 | 274762 |
| total_deaths | 28802 | 33415 | 27127 | 38489 |
| total_cases_pm | 2324.879 | 3853.985 | 5120.952 | 4047.403 |
| total_deaths_pm | 441.251 | 552.663 | 580.197 | 566.965 |
| population | 65273512.0 | 60461828.0 | 46754783.0 | 67886004.0 |
| pop_density | 122.578 | 205.859 | 93.105 | 272.898 |
| median_age | 42.0 | 47.9 | 45.5 | 40.8 |
| gdp_per_capita | 38605.671 | 35220.084 | 34272.36 | 39753.244 |
| hosp_beds | 5.98 | 3.18 | 2.97 | 2.54 |
# 检查回归线下方的意外值
data.loc[(data.total_cases>400000) & (data.total_deaths<10000)].T| iso_code | RUS |
|---|---|
| lastdate | 2020-06-01 |
| location | Russia |
| total_cases | 405843 |
| total_deaths | 4693 |
| total_cases_pm | 2780.995 |
| total_deaths_pm | 32.158 |
| population | 145934460.0 |
| pop_density | 8.823 |
| median_age | 39.6 |
| gdp_per_capita | 24765.954 |
| hosp_beds | 8.05 |
从上表结果来看,有4个国家(法国,意大利,西班牙和英国)的病例数少于30w,死亡人数却超过2w;有1个国家(俄罗斯)的病例数超过30w,但死亡人数少于1w。
3. 使用线性回归来确定具有重大影响的数据点
使用线性回归的优势在于他们对于变量的分布依赖性较小,并且能比单变量或双变量分析揭示更多的东西,识别出在其它方面不显著的异常值。
使用statsmodels OLS方法拟合每百万人口总病例数的线性回归模型,然后确定那些对该模型具有较大影响的国家/地区。
3.1 描述性统计
xvars=['pop_density','median_age','gdp_per_capita']
data_x=data.loc[:,['total_cases_pm']+xvars].dropna()
data_x.describe()| total_cases_pm | pop_density | median_age | gdp_per_capita | |
|---|---|---|---|---|
| count | 175.000000 | 175.000000 | 175.000000 | 175.000000 |
| mean | 1134.015709 | 247.151863 | 30.537143 | 19008.385423 |
| std | 2101.363772 | 822.398967 | 9.117751 | 19673.386571 |
| min | 0.000000 | 1.980000 | 15.100000 | 661.240000 |
| 25% | 67.448000 | 36.066000 | 22.300000 | 4458.202500 |
| 50% | 263.413000 | 82.328000 | 29.700000 | 12951.839000 |
| 75% | 1357.506000 | 207.960000 | 38.700000 | 27467.146000 |
| max | 19753.146000 | 7915.731000 | 48.200000 | 116935.600000 |
3.2 拟合线性回归模型
def getlm(df):
Y=df.total_cases_pm
X=df[['pop_density','median_age','gdp_per_capita']]
X=sm.add_constant(X)
return sm.OLS(Y,X).fit()
lm=getlm(data_x)
lm.summary()| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| const | 944.4731 | 426.712 | 2.213 | 0.028 | 102.172 | 1786.774 |
| pop_density | -0.2057 | 0.142 | -1.447 | 0.150 | -0.486 | 0.075 |
| median_age | -49.4398 | 16.013 | -3.088 | 0.002 | -81.048 | -17.832 |
| gdp_per_capita | 0.0921 | 0.008 | 12.015 | 0.000 | 0.077 | 0.107 |
3.3 确定对模型影响较大的国家/地区,并绘制影响图
在线性回归中,库克距离描述了单个样本对整个回归模型的影响程度,库克距离越大,说明影响越大。当库克距离大于0.5应仔细检查。
influence=lm.get_influence().summary_frame()
influence.loc[influence.cooks_d>0.5,['cooks_d']]| iso_code | cooks_d |
|---|---|
| HKG | 0.780662 |
| QAT | 5.080180 |
data_x.loc[influence.cooks_d>0.5]| total_cases_pm | pop_density | median_age | gdp_per_capita | |
|---|---|---|---|---|
| iso_code | ||||
| HKG | 0.000 | 7039.714 | 44.8 | 56054.92 |
| QAT | 19753.146 | 227.322 | 31.9 | 116935.60 |
fig,ax=plt.subplots(figsize=(12,8))
sm.graphics.influence_plot(lm,ax=ax,criterion="cooks")
plt.show()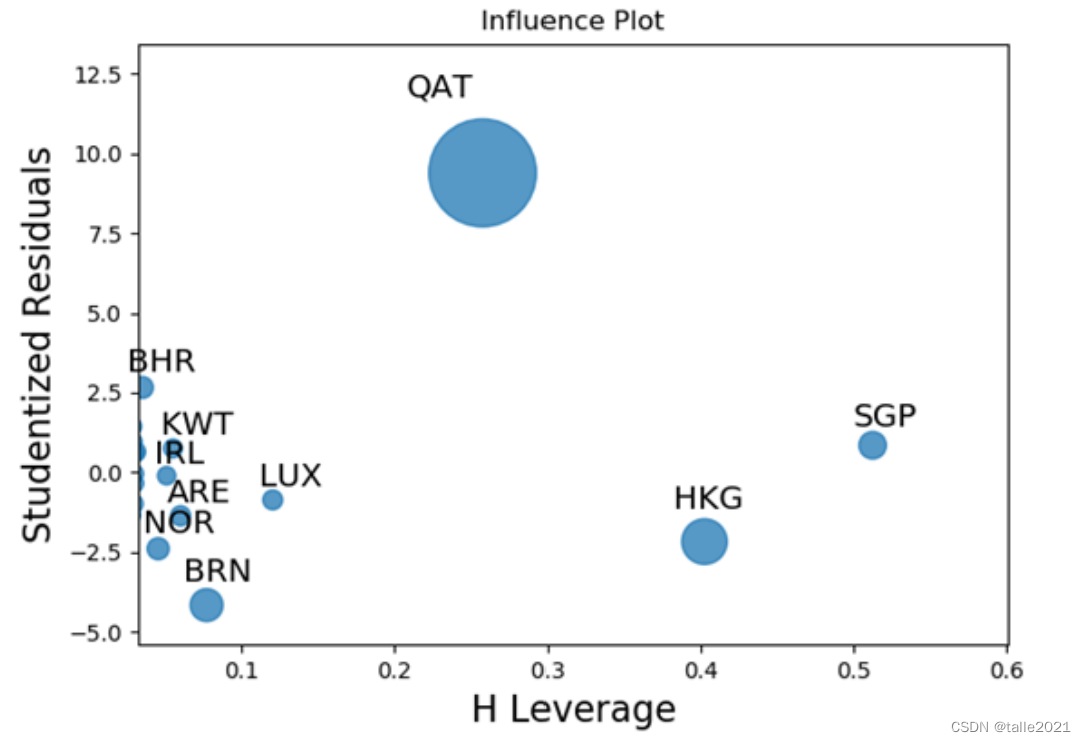
3.4 删除离群点,再进行回归拟合
data_x_new=data_x.loc[influence.cooks_d<0.5]
lm=getlm(data_x_new)
lm.summary()| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| const | 44.0854 | 349.924 | 0.126 | 0.900 | -646.700 | 734.870 |
| pop_density | 0.2423 | 0.145 | 1.666 | 0.098 | -0.045 | 0.529 |
| median_age | -2.5165 | 13.526 | -0.186 | 0.853 | -29.217 | 24.184 |
| gdp_per_capita | 0.0557 | 0.007 | 7.875 | 0.000 | 0.042 | 0.070 |
删除之后,常数和median_age的估计值不再有效。回归输出的P>|t|值表明该系数是否与0有着显著不同。在第一次回归中,median_age和gdp_per_capita的系数在99%的置信水平上是显著的;即P>|t|小于0.01。而在删除两个离群值的情况下,只有gdp_per_capita是显著的。因此在处理此问题时,应分析诸如此类的离群值是否会增加重要信息或使模型失真。
4. 使用k最近邻算法找到离群值
在使用线性回归中,我们使用了每百万人口病例数作为因变量,而在没有标签数据的情况下,即没有目标变量或因变量时,无监督的机器学习工具也可以识别与其它观察结果不同的观察结果。
from pyod.models.knn import KNN
from sklearn.preprocessing import StandardScaler
# 创建分析列的标准化DataFrame
standardizer=StandardScaler()
var1=['location','total_cases_pm', 'total_deaths_pm', 'pop_density','median_age', 'gdp_per_capita']
data_var1=data.loc[:,var1].dropna()
data_var1_stand=standardizer.fit_transform(data_var1.iloc[:,1:])
# KNN模型生成异常分数
clf_name='KNN'
clf=KNN(contamination=0.1) #contamination:异常样本的百分比,在[0,0.5]之间的float型,默认为0.5
clf.fit(data_var1_stand)
y_pred=clf.labels_ # 返回训练数据上的分类标签 (0: 正常值, 1: 异常值)
y_scores=clf.decision_scores_ # 返回训练数据上的异常值 (分值越大越异常)
pred=pd.DataFrame(zip(y_pred,y_scores),columns=['outlier','scores'],index=data_var1.index)pred.outlier.value_counts()0 157 1 18 Name: outlier, dtype: int64
显示离群的数据
data_var1.join(pred).loc[pred.outlier==1,['location','total_cases_pm','total_deaths_pm','scores']].sort_values(['scores'],ascending=False)| location | total_cases_pm | total_deaths_pm | scores | |
|---|---|---|---|---|
| iso_code | ||||
| SGP | Singapore | 5962.727 | 3.931 | 9.228543 |
| HKG | Hong Kong | 0.000 | 0.000 | 7.770444 |
| QAT | Qatar | 19753.146 | 13.190 | 2.643709 |
| BHR | Bahrain | 6698.468 | 11.166 | 2.049642 |
| LUX | Luxembourg | 6418.776 | 175.726 | 1.525783 |
| MDV | Maldives | 3280.041 | 9.250 | 1.395496 |
| MLT | Malta | 1395.120 | 15.854 | 1.353541 |
| BGD | Bangladesh | 286.315 | 3.947 | 1.117904 |
| BRN | Brunei | 322.298 | 4.572 | 1.108011 |
| SAU | Saudi Arabia | 2449.053 | 14.448 | 0.902926 |
| ARE | United Arab Emirates | 3493.994 | 26.693 | 0.822334 |
| KWT | Kuwait | 6332.420 | 49.642 | 0.803449 |
| BRB | Barbados | 320.144 | 24.359 | 0.740434 |
| IRL | Ireland | 5060.962 | 334.562 | 0.721934 |
| PSE | Palestine | 122.907 | 0.980 | 0.717875 |
| NOR | Norway | 1551.489 | 43.532 | 0.712298 |
| GNQ | Equatorial Guinea | 930.872 | 8.553 | 0.695340 |
| OMN | Oman | 2239.641 | 9.204 | 0.670568 |
从上表可以看出,新加坡、卡塔尔的决策分数显著高于其它国家/地区,与前面的分析结果一致。
5. 使用隔离森林算法查找异常
隔离森林(isolation forest,也称孤立森林)找到离群点的方法是,对数据进行连续分区,直至某个数据点被隔离。如果某个数据点仅需要很少的分区即可隔离,那么它将获得较高的异常分数。
from sklearn.ensemble import IsolationForest
from mpl_toolkits.mplot3d import Axes3D
var2=['location','total_cases_pm', 'total_deaths_pm', 'pop_density','median_age', 'gdp_per_capita']
standardizer=StandardScaler()
data_var2=data.loc[:,var2].dropna()
data_var2_stand=standardizer.fit_transform(data_var2.iloc[:,1:])
clf=IsolationForest(n_estimators=100,max_samples='auto',contamination=0.1,max_features=1.0)
clf.fit(data_var2_stand)
data_var2['anomaly']=clf.predict(data_var2_stand) #查看隔离森林得出的结果,输出为一维矩阵,其中值为1代表为正常数据点,值为-1代表为异常点
data_var2['scores']=clf.decision_function(data_var2_stand) #数据集中样本的异常分数值,注意其中值是以0为阈值,小于0视为异常,大于0视为正常,data_var2.anomaly.value_counts()1 157 -1 18 Name: anomaly, dtype: int64
# 创建离群值和内围值的DataFrame
inlier,outlier=data_var2.loc[data_var2.anomaly==1],data_var2.loc[data_var2.anomaly==-1]
outlier[['location','total_cases_pm','total_deaths_pm','anomaly','scores']].sort_values(['scores']).head(10)| location | total_cases_pm | total_deaths_pm | anomaly | scores | |
|---|---|---|---|---|---|
| iso_code | |||||
| QAT | Qatar | 19753.146 | 13.190 | -1 | -0.257773 |
| SGP | Singapore | 5962.727 | 3.931 | -1 | -0.199578 |
| HKG | Hong Kong | 0.000 | 0.000 | -1 | -0.166863 |
| BEL | Belgium | 5037.354 | 816.852 | -1 | -0.116571 |
| BHR | Bahrain | 6698.468 | 11.166 | -1 | -0.110567 |
| LUX | Luxembourg | 6418.776 | 175.726 | -1 | -0.097430 |
| ITA | Italy | 3853.985 | 552.663 | -1 | -0.066504 |
| ESP | Spain | 5120.952 | 580.197 | -1 | -0.060739 |
| IRL | Ireland | 5060.962 | 334.562 | -1 | -0.040109 |
| MDV | Maldives | 3280.041 | 9.250 | -1 | -0.031414 |
ax=plt.axes(projection='3d')
ax.set_title('Isolation Forest Anomaly Detection')
ax.set_zlabel("Cases Per Million")
ax.set_xlabel("GDP Per Capita")
ax.set_ylabel("Median Age")
ax.scatter3D(inlier.gdp_per_capita,inlier.median_age,inlier.total_cases_pm,label="inlier",c='blue')
ax.scatter3D(outlier.gdp_per_capita,outlier.median_age,outlier.total_cases_pm,label="outlier",c='red')
ax.legend()
plt.tight_layout()
plt.show()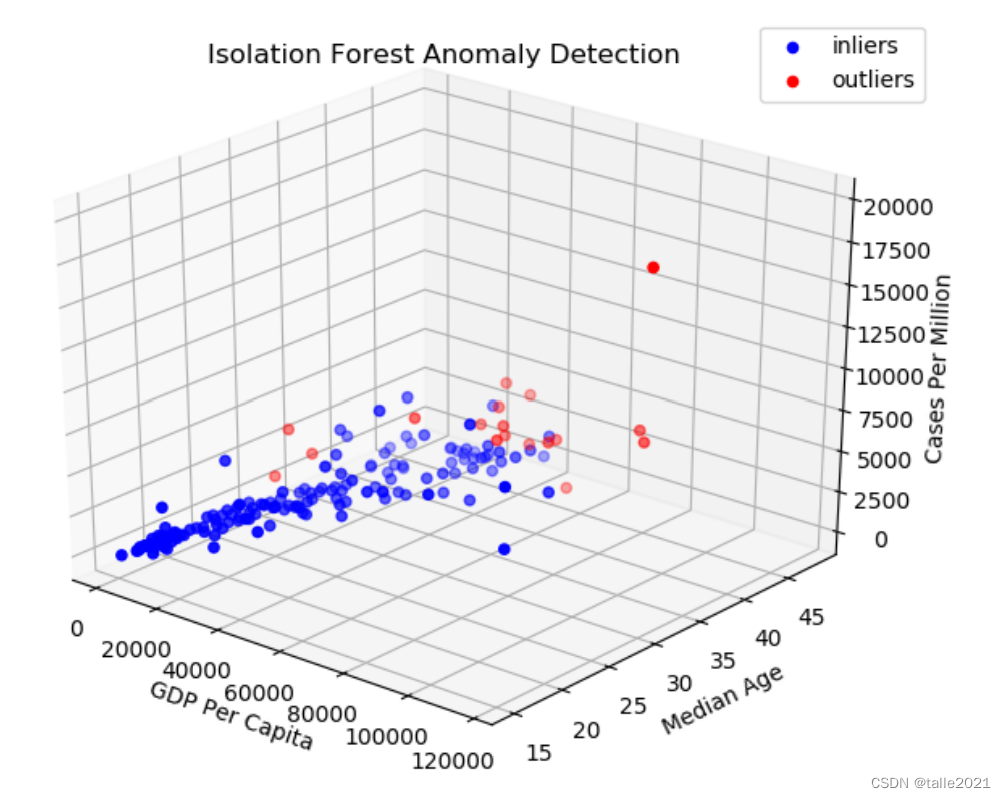
隔离森林的结果与k最近邻的结果非常相似,卡塔尔、新加坡的异常得分较高(准确地说是最小负分),因此,当进行多变量分析时,可以考虑删除离群值再进行分析。