#####安装archr包##别处复制
.libPaths(c("/home/data/t040413/R/x86_64-pc-linux-gnu-library/4.2","/home/data/t040413/R/yll/usr/local/lib/R/site-library", "/usr/local/lib/R/library",
"/home/data/refdir/Rlib/", "/home/data/t040413/R/x86_64-pc-linux-gnu-library/4.2", "/usr/local/lib/R/library"))
.libPaths()
print(getwd())
dir.create("/home/data/t040413/wpx/rna_protein")
setwd("/home/data/t040413/wpx/rna_protein")
#https://www.jianshu.com/p/4b19e7a9bbd3
#https://www.jianshu.com/p/3e5be5e75b20
#https://www.jianshu.com/p/52f462fdb950
#https://www.biostars.org/p/294268/
#https://www.biostars.org/p/287871/
if (F) {
library(DOSE)
#BiocManager::install("GOSemSim")
library(GOSemSim)
library(clusterProfiler)
library(org.Hs.eg.db)
# library(org.Mm.eg.db)
library(org.Rn.eg.db)
library(dplyr)
library(GO.db)
#
get_GO_data <- function(OrgDb, ont, keytype) {
GO_Env <- get_GO_Env()
use_cached <- FALSE
if (exists("organism", envir=GO_Env, inherits=FALSE) &&
exists("keytype", envir=GO_Env, inherits=FALSE)) {
org <- get("organism", envir=GO_Env)
kt <- get("keytype", envir=GO_Env)
if (org == DOSE:::get_organism(OrgDb) &&
keytype == kt &&
exists("goAnno", envir=GO_Env, inherits=FALSE)) {
## https://github.com/GuangchuangYu/clusterProfiler/issues/182
## && exists("GO2TERM", envir=GO_Env, inherits=FALSE)){
use_cached <- TRUE
}
}
if (use_cached) {
goAnno <- get("goAnno", envir=GO_Env)
} else {
OrgDb <- GOSemSim:::load_OrgDb(OrgDb)
kt <- keytypes(OrgDb)
if (! keytype %in% kt) {
stop("keytype is not supported...")
}
kk <- keys(OrgDb, keytype=keytype)
goAnno <- suppressMessages(
AnnotationDbi::select(OrgDb, keys=kk, keytype=keytype,
columns=c("GOALL", "ONTOLOGYALL")))
goAnno <- unique(goAnno[!is.na(goAnno$GOALL), ])
assign("goAnno", goAnno, envir=GO_Env)
assign("keytype", keytype, envir=GO_Env)
assign("organism", DOSE:::get_organism(OrgDb), envir=GO_Env)
}
if (ont == "ALL") {
GO2GENE <- unique(goAnno[, c(2,1)])
} else {
GO2GENE <- unique(goAnno[goAnno$ONTOLOGYALL == ont, c(2,1)])
}
GO_DATA <- DOSE:::build_Anno(GO2GENE, get_GO2TERM_table())
goOnt.df <- goAnno[, c("GOALL", "ONTOLOGYALL")] %>% unique
goOnt <- goOnt.df[,2]
names(goOnt) <- goOnt.df[,1]
assign("GO2ONT", goOnt, envir=GO_DATA)
return(GO_DATA)
}
get_GO_Env <- function () {
if (!exists(".GO_clusterProfiler_Env", envir = .GlobalEnv)) {
pos <- 1
envir <- as.environment(pos)
assign(".GO_clusterProfiler_Env", new.env(), envir=envir)
}
get(".GO_clusterProfiler_Env", envir = .GlobalEnv)
}
get_GO2TERM_table <- function() {
GOTERM.df <- get_GOTERM()
GOTERM.df[, c("go_id", "Term")] %>% unique
}
get_GOTERM <- function() {
pos <- 1
envir <- as.environment(pos)
if (!exists(".GOTERM_Env", envir=envir)) {
assign(".GOTERM_Env", new.env(), envir)
}
GOTERM_Env <- get(".GOTERM_Env", envir = envir)
if (exists("GOTERM.df", envir = GOTERM_Env)) {
GOTERM.df <- get("GOTERM.df", envir=GOTERM_Env)
} else {
GOTERM.df <- toTable(GOTERM)
assign("GOTERM.df", GOTERM.df, envir = GOTERM_Env)
}
return(GOTERM.df)
}
print(getwd())
dir.create("/home/data/t040413/wpx/rna_protein")
setwd("/home/data/t040413/wpx/rna_protein")
}
library(GO.db)
ls("package:GO.db")
columns(GO.db)
GO_dbInfo()
GO_dbconn()
GO_dbfile()
# Find the children terms of the GO term "GO:0006412"
children_terms <- as.list(GOBPCHILDREN["GO:0006412"])
as.list(GOBPCHILDREN["GO:0006412"])
as.list(GOBPOFFSPRING["GO:0006412"])
if (F) {
# Print the children terms
print(children_terms)
## Bimap interface:
# Convert the object to a list
xx <- as.list(GOBPOFFSPRING)
xx
# Remove GO IDs that do not have any offspring
xx <- xx[!is.na(xx)]
if(length(xx) > 0){
# Get the offspring GO IDs for the first two elents of xx
goids <- xx[1:2]
}
## Bimap interface:
# Convert the object to a list
xx <- as.list(GOBPANCESTOR)
# Remove GO IDs that do not have any ancestor
xx <- xx[!is.na(xx)]
if(length(xx) > 0){
# Get the ancestor GO IDs for the first two elents of xx
goids <- xx[1:2]
}
goids
# Convert the object to a list
xx <- as.list(GOBPCHILDREN)
# Remove GO IDs that do not have any children
xx <- xx[!is.na(xx)]
xx
if(length(xx) > 0){
# Get the parent GO IDs for the first elents of xx
goids <- xx[[1]]
# Find out the GO terms for the first parent goid
GOID(GOTERM[[goids[1]]])
Term(GOTERM[[goids[1]]])
Synonym(GOTERM[[goids[1]]])
Secondary(GOTERM[[goids[1]]])
Definition(GOTERM[[goids[1]]])
Ontology(GOTERM[[goids[1]]])
}
goids
}
myterm= c("GO:0007612")
union(as.list(GOBPOFFSPRING[myterm]) %>%unlist() %>% unname(), names(as.list(GOBPOFFSPRING[myterm])))
library(AnnotationDbi)
k <- keys(GO.db, keytype = "GOID")[1:3]
AnnotationDbi::select(GO.db,
keys = k,
columns = c("TERM","ONTOLOGY"),
keytype="GOID")
print(getwd())#----------------------------==========================================-==================================
load("/home/data/t040413/wpx/rna_protein/selected_go_terms.rds")
# go_file=list.files("~/wpx/wpx_transcriptomics_proteinomics_Metabolomics/transcriptomics/4_d28_vs_d14_treatment_effective_2",
# full.names = TRUE,pattern = "GO_enrichment.xlsx")
go_file=list.files(".",full.names = TRUE,pattern = "GO_enrichment.xlsx")[1]
a
rna_msc=openxlsx::read.xlsx(go_file)
head(rna_msc)
rna_msc=rna_msc[grepl(rna_msc$Cluster,pattern="MSC-D28 VS LCT\\(14\\+28\\)-NT"),]
head(rna_msc)
rna_msc
rna_msc_id=rna_msc$ID
a
if (F) {
as.list(GOBPOFFSPRING[myterm]) %>%unlist() %>% unname()
as.list(GOBPOFFSPRING['GO:0016209']) %>%unlist() %>% unname()
for (eachterm in a) {
result <- try({
try( as.list(GOBPOFFSPRING[eachterm]) %>% unlist() %>% unname() %>%print() , silent = TRUE ) |
try(as.list(GOMFOFFSPRING[eachterm]) %>% unlist() %>% unname()%>%print(), silent = TRUE)
}, silent = TRUE)
print(paste0(eachterm,"-====================================================================="))
# # if (inherits(result, "try-error")) {
# cat("Error occurred for term:", eachterm, "\n")
# }
}
#grep(as.list(GOBPOFFSPRING) %>%unlist() %>%unname(),pattern("0016209"))
rna_msc_match=list()
if (F) {
for (eachterm in a) {
success <- TRUE
tryCatch({
target_terms <- union(as.list(GOBPOFFSPRING[eachterm]) %>% unlist() %>% unname(), names(as.list(GOBPOFFSPRING[eachterm])))
rna_msc_match[[eachterm]] <- rna_msc[rna_msc_id %in% target_terms,]
print(paste0(eachterm, "-========================GO============================================="))
}, error = function(e) {#因为这是函数体内的函数 所以rna_msc_match与体外无关,如何改进呢
cat("Error occurred for term:", eachterm, "\n")
success <- FALSE
print(paste(success,"-===="))
if (!success) {
target_terms <- union(as.list(GOMFOFFSPRING[eachterm]) %>% unlist() %>% unname(), names(as.list(GOMFOFFSPRING[eachterm])))
rna_msc_match[[eachterm]] <- rna_msc[rna_msc_id %in% target_terms,]
print(paste0(eachterm,100-1, "-===============================MF======================================"))
}
})
}
a
names(rna_msc_match)
}
}
colnames(GOBPOFFSPRING)
as.list(GOBPOFFSPRING)
rna_msc_match <- list()
i=1
for (eachterm in a) {
# Check if GOBPOFFSPRING has the term, otherwise use GOMFOFFSPRING
if ( eachterm %in% (union(as.list(GOBPOFFSPRING[[eachterm]]) %>% unlist() %>% unname(), eachterm)) ) {
target_terms <- union(as.list(GOBPOFFSPRING[[eachterm]]) %>% unlist() %>% unname(), eachterm)
print(paste0(eachterm,"___第几个::",i, "-========================GO============================================="))
} else {
target_terms <- union(as.list(GOMFOFFSPRING[[eachterm]]) %>% unlist() %>% unname(), eachterm)
print(paste0(eachterm,"___第几个::",i, "-========================MF============================================="))
}
# Filter rna_msc based on target_terms
rna_msc_match[[eachterm]] <- rna_msc[rna_msc_id %in% target_terms,]
i=i+1
}
a
rna_msc_match %>%names()
rna_msc_match
a_list=rna_msc_match
do.call(cbind, lapply(lapply(a_list, unlist), `length<-`, max(lengths(a_list))))
# cat months dog
my_list=a_list
# Step 1: 合并数据框
combined_df <- do.call(rbind, my_list)
# Step 2: 添加列表名
combined_df$List_Name <- unlist(lapply(seq_along(my_list), function(i) rep(names(my_list)[i], nrow(my_list[[i]]))))
a
combined_df=combined_df[!duplicated(combined_df$Description),]
combined_df
combined_df$Enrichment= -log2(combined_df$p.adjust) %>%as.numeric()
combined_df$"-log2(combined_df$pvalue)"=-log2(combined_df$pvalue)
combined_df$"-log2(combined_df$p.adjust)"=-log2(combined_df$p.adjust)
combined_df$Percentage=DOSE::parse_ratio(combined_df$GeneRatio) /DOSE::parse_ratio(combined_df$BgRatio)
combined_df
#library(tibble)
terms_for_heatmap=data.frame(row.names =combined_df$Description,
Enrichment=combined_df$Enrichment)
terms_for_heatmap
pheatmap(terms_for_heatmap,cluster_cols = FALSE,
scale = "none",
color = colorRampPalette(c( "pink","red"))(50),
cluster_rows = FALSE,gaps_row = seq(1,length(rownames(terms_for_heatmap)))
)
#蛋白组
if (1) {
go_file=list.files(".",full.names = TRUE,pattern = "protein")
a
go_file
protein_msc=openxlsx::read.xlsx(go_file)
head(protein_msc)
protein_msc=protein_msc[grepl(protein_msc$Cluster,pattern="MSC_D28-LCT_14_28_NT_D28"),]
head(protein_msc)
protein_msc
protein_msc_id=protein_msc$ID
a
if (F) {
as.list(GOBPOFFSPRING[myterm]) %>%unlist() %>% unname()
as.list(GOBPOFFSPRING['GO:0016209']) %>%unlist() %>% unname()
for (eachterm in a) {
result <- try({
try( as.list(GOBPOFFSPRING[eachterm]) %>% unlist() %>% unname() %>%print() , silent = TRUE ) |
try(as.list(GOMFOFFSPRING[eachterm]) %>% unlist() %>% unname()%>%print(), silent = TRUE)
}, silent = TRUE)
print(paste0(eachterm,"-====================================================================="))
# # if (inherits(result, "try-error")) {
# cat("Error occurred for term:", eachterm, "\n")
# }
}
#grep(as.list(GOBPOFFSPRING) %>%unlist() %>%unname(),pattern("0016209"))
protein_msc_match=list()
if (F) {
for (eachterm in a) {
success <- TRUE
tryCatch({
target_terms <- union(as.list(GOBPOFFSPRING[eachterm]) %>% unlist() %>% unname(), names(as.list(GOBPOFFSPRING[eachterm])))
protein_msc_match[[eachterm]] <- protein_msc[protein_msc_id %in% target_terms,]
print(paste0(eachterm, "-========================GO============================================="))
}, error = function(e) {#因为这是函数体内的函数 所以protein_msc_match与体外无关,如何改进呢
cat("Error occurred for term:", eachterm, "\n")
success <- FALSE
print(paste(success,"-===="))
if (!success) {
target_terms <- union(as.list(GOMFOFFSPRING[eachterm]) %>% unlist() %>% unname(), names(as.list(GOMFOFFSPRING[eachterm])))
protein_msc_match[[eachterm]] <- protein_msc[protein_msc_id %in% target_terms,]
print(paste0(eachterm,100-1, "-===============================MF======================================"))
}
})
}
a
names(protein_msc_match)
}
}
colnames(GOBPOFFSPRING)
as.list(GOBPOFFSPRING)
protein_msc_match <- list()
i=1
for (eachterm in a) {
# Check if GOBPOFFSPRING has the term, otherwise use GOMFOFFSPRING
if ( eachterm %in% (union(as.list(GOBPOFFSPRING[[eachterm]]) %>% unlist() %>% unname(), eachterm)) ) {
target_terms <- union(as.list(GOBPOFFSPRING[[eachterm]]) %>% unlist() %>% unname(), eachterm)
print(paste0(eachterm,"___第几个::",i, "-========================GO============================================="))
} else {
target_terms <- union(as.list(GOMFOFFSPRING[[eachterm]]) %>% unlist() %>% unname(), eachterm)
print(paste0(eachterm,"___第几个::",i, "-========================MF============================================="))
}
# Filter protein_msc based on target_terms
protein_msc_match[[eachterm]] <- protein_msc[protein_msc_id %in% target_terms,]
i=i+1
}
a
protein_msc_match %>%names()
protein_msc_match
a_list=protein_msc_match
do.call(cbind, lapply(lapply(a_list, unlist), `length<-`, max(lengths(a_list))))
# cat months dog
my_list=a_list
# Step 1: 合并数据框
combined_df_protein <- do.call(rbind, my_list)
# Step 2: 添加列表名
combined_df_protein$List_Name <- unlist(lapply(seq_along(my_list), function(i) rep(names(my_list)[i], nrow(my_list[[i]]))))
a
combined_df_protein=combined_df_protein[!duplicated(combined_df_protein$Description),]
combined_df_protein
combined_df_protein$Enrichment= -log2(combined_df_protein$p.adjust) %>%as.numeric()
combined_df_protein$"-log2(combined_df_protein$pvalue)"=-log2(combined_df_protein$pvalue)
combined_df_protein$"-log2(combined_df_protein$p.adjust)"=-log2(combined_df_protein$p.adjust)
combined_df_protein$Percentage=DOSE::parse_ratio(combined_df_protein$GeneRatio) /DOSE::parse_ratio(combined_df_protein$BgRatio)
combined_df_protein
#library(tibble)
terms_for_heatmap_protein=data.frame(row.names =combined_df_protein$Description,
Enrichment=combined_df_protein$Enrichment)
terms_for_heatmap_protein
pheatmap(terms_for_heatmap_protein,cluster_cols = FALSE,
scale = 'none',
color = colorRampPalette(c( "pink","red"))(50),
cluster_rows = FALSE,gaps_row = seq(1,length(rownames(terms_for_heatmap_protein)))
)
}
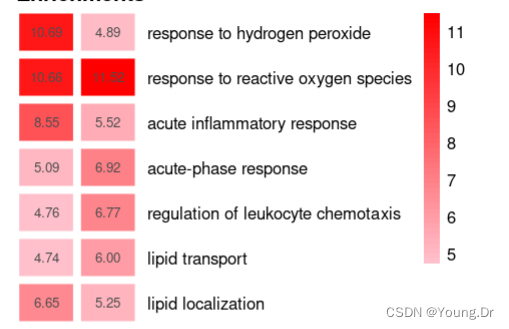
common=intersect(rownames(terms_for_heatmap),rownames(terms_for_heatmap_protein))
common_df=data.frame(transcript_enrichment=terms_for_heatmap[common,],
protein_enrichment=terms_for_heatmap_protein[common,],
names=common)
list_save=list(transcript=combined_df,
protein=combined_df_protein,
combined=common_df)
openxlsx::write.xlsx(list_save,file = "commbined_protein_transcript_GO_enrichments.xlsx")
library(ggplot2)
library(reshape2)
data=data.frame(transcript_enrichment=terms_for_heatmap[common,],
protein_enrichment=terms_for_heatmap_protein[common,],
row.names =common)
combined_df %>%head()
p=pheatmap::pheatmap(data,
cellwidth = 33,
cellheight = 23,border_color = "white",
legend_labels = "Enrichments",
main = "Enrichments",
cluster_cols = FALSE,
scale = 'none',
color = colorRampPalette(c( "pink","red"))(50),
cluster_rows = FALSE,gaps_row = seq(1,length(rownames(data))),
gaps_col = seq(1,length(colnames(data)))
)
ggsave(plot = p,filename = "commbined_protein_transcript_GO_enrichment.pdf",
height = 10,width = 10,
limitsize = FALSE)
p=pheatmap::pheatmap(data,
cellwidth = 33,
cellheight = 23,border_color = "white",
legend_labels = "Enrichments",
main = "Enrichments",display_numbers = T,
cluster_cols = FALSE,
scale = 'none',
color = colorRampPalette(c( "pink","red"))(50),
cluster_rows = FALSE,gaps_row = seq(1,length(rownames(data))),
gaps_col = seq(1,length(colnames(data)))
)
ggsave(plot = p,filename = "commbined_protein_transcript_GO_enrichment_with_numbers.pdf",
height = 10,width = 10,
limitsize = FALSE)
