2.5.1. 主成分分析(PCA)¶
2.5.1.1. 准确的PCA和概率解释(Exact PCA and probabilistic interpretation)
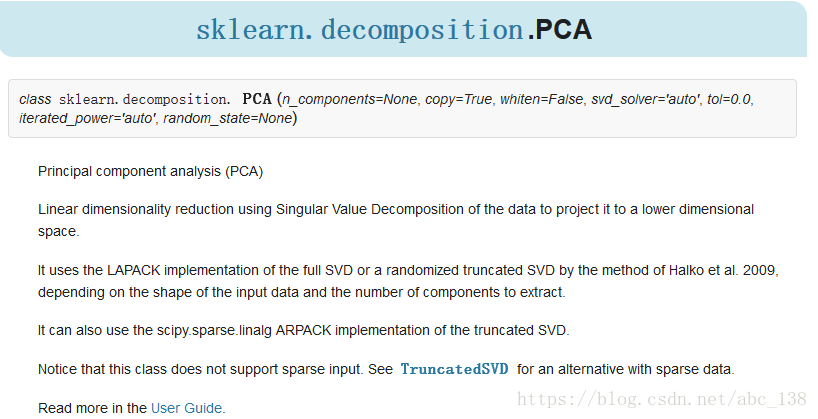
PCA 用于对一组连续正交分量中的多变量数据集进行方差最大方向的分解。在 scikit-learn 中, PCA 被实现为一个变换对象, 通过 fit 方法可以降维成 n 个成分,并且可以将新的数据投影(project, 亦可理解为分解)到这些成分中。
可选参数 whiten=True 使得可以将数据投影到奇异(singular)空间上,同时将每个成分缩放到单位方差。如果下游模型对信号的各向同性作出强烈的假设,这通常是有用的,例如,使用RBF内核的 SVM 算法和 K-Means 聚类算法。
PCA 对象还提供了 PCA 的概率解释, 其可以基于其解释的方差量给出数据的可能性。
可以通过在交叉验证(cross-validation)中使用 score 方法来实现:
一 下面对主要的参数进行解释
Parameters:
n_components : int, float, None or string
Number of components to keep.if n_components is not set all components are kept:
n_components == min(n_samples, n_features)if n_components == ‘mle’ and svd_solver == ‘full’, Minka’s MLE is usedto guess the dimensionif
0 < n_components < 1and svd_solver == ‘full’, select the numberof components such that the amount of variance that needs to beexplained is greater than the percentage specified by n_componentsn_components cannot be equal to n_features for svd_solver == ‘arpack’.
copy : bool (default True)
If False, data passed to fit are overwritten and runningfit(X).transform(X) will not yield the expected results,use fit_transform(X) instead.
whiten : bool, optional (default False)
When True (False by default) the components_ vectors are multipliedby the square root of n_samples and then divided by the singular valuesto ensure uncorrelated outputs with unit component-wise variances.
Whitening will remove some information from the transformed signal(the relative variance scales of the components) but can sometimeimprove the predictive accuracy of the downstream estimators bymaking their data respect some hard-wired assumptions.
svd_solver : string {‘auto’, ‘full’, ‘arpack’, ‘randomized’}
- auto :
the solver is selected by a default policy based on X.shape andn_components: if the input data is larger than 500x500 and thenumber of components to extract is lower than 80% of the smallestdimension of the data, then the more efficient ‘randomized’method is enabled. Otherwise the exact full SVD is computed andoptionally truncated afterwards.
- full :
run exact full SVD calling the standard LAPACK solver viascipy.linalg.svd and select the components by postprocessing
- arpack :
run SVD truncated to n_components calling ARPACK solver viascipy.sparse.linalg.svds. It requires strictly0 < n_components < X.shape[1]
- randomized :
run randomized SVD by the method of Halko et al.
tol : float >= 0, optional (default .0)
Tolerance for singular values computed by svd_solver == ‘arpack’.
iterated_power : int >= 0, or ‘auto’, (default ‘auto’)
Number of iterations for the power method computed bysvd_solver == ‘randomized’.
random_state : int, RandomState instance or None, optional (default None)
If int, random_state is the seed used by the random number generator;If RandomState instance, random_state is the random number generator;If None, the random number generator is the RandomState instance usedby np.random. Used when svd_solver == ‘arpack’ or ‘randomized’.
(1)n_components:int,float,None或string
要保留的主成分数量。如果未设置n_components,则保留所有主成分:
n_components == min(n_samples,n_features)
如果n_components =='mle'和svd_solver =='full',则使用Minka的MLE来猜测维度,如果0 <n_components <1和svd_solver =='full',选择组件的数量,使得需要的变化量需要说明的是大于由n_components指定的百分比n_components不能等于n_features对于svd_solver =='arpack'。
(2)copy:bool(默认为True)
如果为False,则传递给拟合的数据将被覆盖并运行拟合(X).transform(X)将不会产生预期结果,请改为使用fit_transform(X)。
(3)whiten :判断是否进行白化。所谓白化,就是对降维后的数据的每个特征进行归一化,让方差都为1.对于PCA降维本身来说,一般不需要白化。如果你PCA降维后有后续的数据处理动作,可以考虑白化。默认值是False,即不进行白化。
(4)svd_solver:即指定奇异值分解SVD的方法,由于特征分解是奇异值分解SVD的一个特例,一般的PCA库都是基于SVD实现的。有4个可以选择的值:{‘auto’, ‘full’, ‘arpack’, ‘randomized’}。randomized一般适用于数据量大,数据维度多同时主成分数目比例又较低的PCA降维,它使用了一些加快SVD的随机算法。 full则是传统意义上的SVD,使用了scipy库对应的实现。arpack和randomized的适用场景类似,区别是randomized使用的是scikit-learn自己的SVD实现,而arpack直接使用了scipy库的sparse SVD实现。默认是auto,即PCA类会自己去在前面讲到的三种算法里面去权衡,选择一个合适的SVD算法来降维。一般来说,使用默认值就够了。
二 对象属性
Attributes:
components_ : array, shape (n_components, n_features)
Principal axes in feature space, representing the directions ofmaximum variance in the data. The components are sorted by
explained_variance_.
explained_variance_ : array, shape (n_components,)
The amount of variance explained by each of the selected components. Equal to n_components largest eigenvaluesof the covariance matrix of X.
explained_variance_ratio_ : array, shape (n_components,)
Percentage of variance explained by each of the selected components. If
n_componentsis not set then all components are stored and thesum of explained variances is equal to 1.0.
singular_values_ : array, shape (n_components,)
The singular values corresponding to each of the selected components.The singular values are equal to the 2-norms of the
n_componentsvariables in the lower-dimensional space.(1)components_:数组,形状(n_components,n_features)
特征空间中的主轴,代表数据中最大方差的方向。 这些组件按descriptions_variance_排序。(2)explained_variance_,它代表降维后的各主成分的方差值。方差值越大,则说明越是重要的主成分。
(3)explained_variance_ratio_,它代表降维后的各主成分的方差值占总方差值的比例,这个比例越大,则越是重要的主成分。
三 PCA对象的方法
Methods
fit(X[, y]) |
Fit the model with X. |
fit_transform(X[, y]) |
Fit the model with X and apply the dimensionality reduction on X. |
get_covariance() |
Compute data covariance with the generative model. |
get_params([deep]) |
Get parameters for this estimator. |
get_precision() |
Compute data precision matrix with the generative model. |
inverse_transform(X) |
Transform data back to its original space. |
score(X[, y]) |
Return the average log-likelihood of all samples. |
score_samples(X) |
Return the log-likelihood of each sample. |
set_params(**params) |
Set the parameters of this estimator. |
transform(X) |
Apply dimensionality reduction to X. |
fit
(
X,
y=None
)
¶ 训练模型-
Fit the model with X.
Parameters: X : array-like, shape (n_samples, n_features)
Training data, where n_samples in the number of samplesand n_features is the number of features.
Returns: self : object
Returns the instance itself.
fit_transform
(
X,
y=None
) 训练并降维-
Fit the model with X and apply the dimensionality reduction on X.
Parameters: X : array-like, shape (n_samples, n_features)
Training data, where n_samples is the number of samplesand n_features is the number of features.
Returns: X_new : array-like, shape (n_samples, n_components)
fit_transform(X)
用X来训练PCA模型,同时返回降维后的数据。
newX=pca.fit_transform(X),newX就是降维后的数据。
(3)transform(X)¶ 执行降维
-
Apply dimensionality reduction to X.
X is projected on the first principal components previously extractedfrom a training set.
Parameters: X : array-like, shape (n_samples, n_features)
New data, where n_samples is the number of samplesand n_features is the number of features.
Returns: X_new : array-like, shape (n_samples, n_components)
将维度降低应用于X.
将X投影到先前从训练集中提取的第一个主成分上。
参数:
X:类似数组的形状(n_samples,n_features)
新数据,其中n_samples是样本的数量,n_features是要素的数量。
返回:
X_new:类似数组的形状(n_samples,n_components)