1.原始数据:
原始数据链接:
http://current.geneontology.org/ontology/go-basic.obo
原始数据样式:
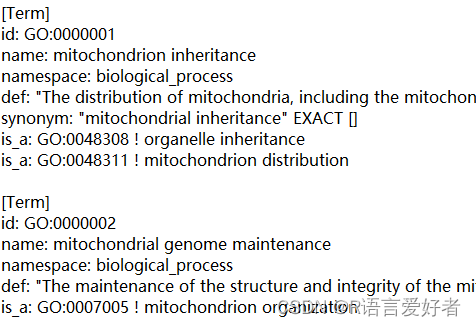
2.目标格式:
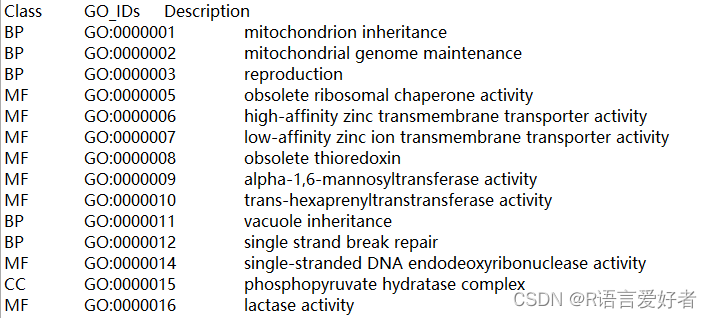
3.代码:
with open("go-basic.obo","r") as file:
lib={
}
for line in file:
line=line.strip()
col_name=line.split(":")[0]
if col_name == "id":
id=line.split(" ",maxsplit=1)[1]
lib[id]=""
if col_name == "name":
name=line.split(" ",maxsplit=1)[1]
lib[id]=lib[id]+"@"+name
if col_name == "namespace":
namespace=line.split(" ",maxsplit=1)[1]
lib[id]=lib[id]+"@"+namespace
out=open("GO_basic_Description.txt","a+")
out.write("Class"+"\t"+"GO_IDs"+"\t"+"Description"+"\n")
for key in lib.keys():
go_id=key
go_name=lib[key].split("@")[1]
go_namespace=lib[key].split("@")[2]
if go_namespace == "molecular_function":
go_namespace="MF"
out.write(go_namespace+"\t"+go_id+"\t"+go_name+"\n")
if go_namespace == "biological_process":
go_namespace="BP"
out.write(go_namespace+"\t"+go_id+"\t"+go_name+"\n")
if go_namespace == "cellular_component":
go_namespace="CC"
out.write(go_namespace+"\t"+go_id+"\t"+go_name+"\n")