sklearn.metrics.confusion_matrix(y_true, y_pred, labels=None, sample_weight=None)
虽然sklearn.metrics.confusion_matrix很方便可以直接得到混淆矩阵,但是由于我实践后发现问题:sklearn的混淆矩阵实现会在只有一类的时候自动降维成1*1,于是我自己手撸一个:
def calculate_metric(gt, pred):
pred[pred>0.5]=1
pred[pred<1]=0
TP, FP, TN, FN = 0, 0, 0, 0
for i in range(len(gt)):
if gt[i] == 1 and pred[i] == 1:
TP += 1
if gt[i] == 0 and pred[i] == 1:
FP += 1
if gt[i] == 0 and pred[i] == 0:
TN += 1
if gt[i] == 1 and pred[i] == 0:
FN += 1
# confusion = confusion_matrix(gt,pred)
# print(confusion.shape)
# TP = confusion[1,1]
# TN = confusion[0,0]
# FP = confusion[0,1]
# FN = confusion[1,0]
return TP, FP, TN, FN但后来发现问题怪不到confusion_matrix,其实是因为数据样本较少,从而导致五折交叉验证时每一折不能保证每一折都包含两类样本,万幸:
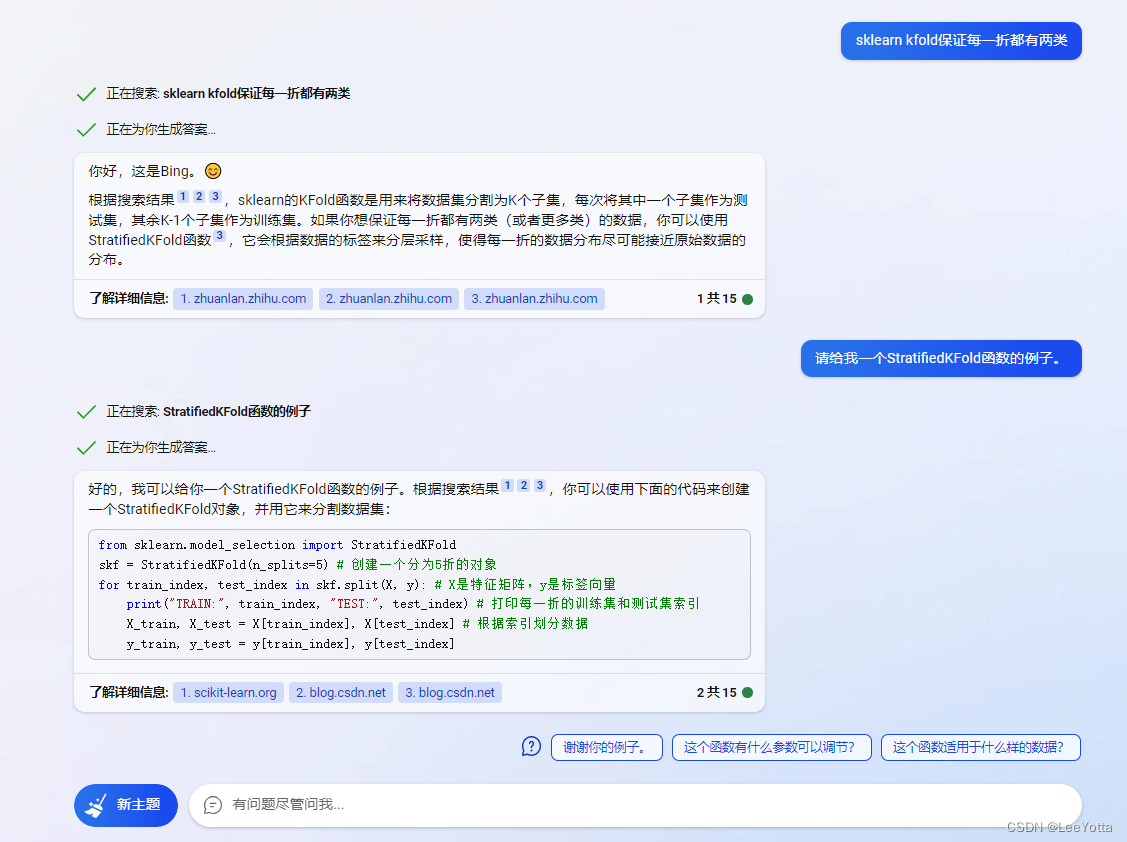
所以去了解了下KFold与StratifiedKFold,两者用法一样,只需要修改函数名就好:
from sklearn.model_selection import train_test_split, KFold, StratifiedKFold
kf = KFold(n_splits=5,random_state=2023,shuffle=True)
kf = StratifiedKFold(n_splits=5,random_state=2023,shuffle=True)
但在做split时两者有部分不同
#KFold不需要传入标签
for train_index, validate_index in kf.split(dataset):
pass
#StratifiedKFold需要传入标签
for train_index, validate_index in kf.split(dataset,dataset['label']):
pass换成StratifiedKFold分层采样就不会有一折只有一类的情况,从而也避免了confusion_matrix的降维问题,至此问题解决。