scala机器学习之保险数据预测分析
数据资料来源:
此文章数据内容来源于Scala Machine Learning Projects 2018版 一书,本书分为是一个章节,本文章内容来自于第一章节
书本链接:Scala Machine Learning Projects
文章所用数据链接:百度云盘链接:提取码:m4ma
代码中出现的参数解释:sparkML各种参数解释
前言
由于该书目前没有中文的翻译文档,所以该书的很多地方是根据个人的理解进行的翻译,个人英语水平不是很好,希望看这篇文章的大佬们多多体谅。
先大致说一下这边文章的主要内容,本文是作者在自学机器学习时,自己的经验分享,仅供大家参考交流使用。Scala Machine Learning Projects这本书共十一个章节,笔者也会慢慢随着自己的学习进度不断更新。此次实战主要内容为该书的第一章节:保险数据的预测分析,本章节通过对比分析三种机器学习算法(主要为:LR,GBT,RF)对于同一数据的预测结果,对比分析不同机器学习算法的模型性能。下面进入正题。
算法简介
LR
本人单独用了一篇文章来叙述LR算法的原理和简易的代码实现,感兴趣的可以点击下面的链接,这里由于篇幅有限,不在赘述。
GBT
RF
代码
SparkSession 初始化
object SparkSessionCreate {
/**
* init SparkSession
* @return SparkSession
*/
def createSession(): SparkSession = {
val spark = SparkSession.builder().master("local[3]").appName("MachineLearn.ScalaMl").getOrCreate()
return spark
}
}
训练数据和测试数据准备与前期过滤
object Preproessing {
val train = "file:\\C:\\Users\\PC\\Desktop\\墨菲斯文件备份\\Word文档\\学习资料\\spark\\书\\机器学习\\数据\\train.csv"
val test = "file:\\C:\\Users\\PC\\Desktop\\墨菲斯文件备份\\Word文档\\学习资料\\spark\\书\\机器学习\\数据\\test.csv"
var trainSample = 1.0
var testSample = 1.0
//获取数据
val ss: SparkSession = SparkSessionCreate.createSession()
val testInput = getDataFromPath(ss, test)
val trainInput = getDataFromPath(ss, train)
//将测试数据分成两部分,比例为0.75/0.25,分别为测试数据和交叉验证数据
var data: Dataset[Row] = trainInput.withColumnRenamed("loss", "label").sample(false, trainSample)
//检查数据中是否存在null值
var DF: DataFrame = data.na.drop()
if (data == DF)
println("DF中没有null值")
else {
data = DF
println("DF中存在null值")
}
val seed = 12345L
//切分数据
val splits: Array[Dataset[Row]] = data.randomSplit(Array(0.75, 0.25), seed)
//将数据分为训练数据和验证数据两部分
val (trainData, validationData): (Dataset[Row], Dataset[Row]) = (splits(0), splits(1))
//cache数据
trainData.cache()
validationData.cache()
//从测试数据中取样
val testData: Dataset[Row] = testInput.sample(false, testSample).cache()
//数据中包含数值和分类值,我们需要区分出来
val featureCols: Array[String] = trainData
.columns
.filter(removeTooManyCategs)
.filter(onlyFeatureCols)
.map(categNewCol)
//将字符串列的标签通过StringIndexer进行标签索引化
val stringIndexerStages: Array[StringIndexerModel] = trainData.columns.filter(isCateg).map(C => new StringIndexer()
.setInputCol(C)
.setOutputCol(categNewCol(C))
.fit(trainInput.select(C).union(testInput.select(C)))
)
val assembler = new VectorAssembler()
.setInputCols(featureCols)
.setOutputCol("features")
def isCateg(C: String): Boolean = C.startsWith("cat")
def categNewCol(C: String): String = if (isCateg(C)) s"idx_${C}" else C
def removeTooManyCategs(C: String): Boolean = !(C matches
"cat(109$|110$|112$|113$|116$)")
def onlyFeatureCols(C: String): Boolean = !(C matches "id|label")
def getDataFromPath(ss: SparkSession, path: String): DataFrame = {
val data: DataFrame = ss.read.option("header", "true").option("inferSchema", "true").format("com.databricks.spark.csv").load(path).cache
return data
}
}
LR算法代码
object ScalaLR {
def main(args: Array[String]): Unit = {
val ss: SparkSession = SparkSessionCreate.createSession()
import ss.implicits._
//定义参数
val numFolds = 10
val MaxIter: Seq[Int] = Seq(1000)
val Regparam: Seq[Double] = Seq(0.001)
val Tol: Seq[Double] = Seq(1e-6)
val ElasticNetParam: Seq[Double] = Seq(0.001)
//创建一个LR估量模型
val model = new LinearRegression()
.setFeaturesCol("features")
.setLabelCol("label")
//创建一个ML pipeline
val pipeline = new Pipeline()
.setStages((Preproessing.stringIndexerStages :+ Preproessing.assembler) :+ model)
//进行交叉验证之前,我们需要指定一些验证参数,下面创建一个paramGrid来指定参数设置
val paramGrid: Array[ParamMap] = new ParamGridBuilder()
.addGrid(model.maxIter, MaxIter)
.addGrid(model.regParam, Regparam)
.addGrid(model.tol, Tol)
.addGrid(model.elasticNetParam, ElasticNetParam)
.build()
//为了更好的交叉验证性能,进行模型调优,参数自行设置,这里的参数为 numFolds = 10
val cv: CrossValidator = new CrossValidator()
.setEstimator(pipeline)
.setEvaluator(new RegressionEvaluator())
.setEstimatorParamMaps(paramGrid)
.setNumFolds(numFolds)
//创建了交叉验证模型器之后,我们可以来训练这个LR模型了
val cvModel = cv.fit(Preproessing.trainData)
//现在我们有了fit模型后,就可以做一些predict行为了,现在,我们可以在这个模型上用train数据和test数据进行模型评估了
val trainPredictionsAndLabels: RDD[(Double, Double)] = cvModel
.transform(Preproessing.trainData)
.select("label", "prediction")
.map { case Row(label: Double, prediction: Double) => (label, prediction) }.rdd
val validPredictionsAndLabels: RDD[(Double, Double)] = cvModel
.transform(Preproessing.validationData)
.select("label", "prediction")
.map { case Row(label: Double, prediction: Double) => (label, prediction) }.rdd
val trainRegressionMetrics = new RegressionMetrics(trainPredictionsAndLabels)
val validRegressionMetrics: RegressionMetrics = new RegressionMetrics(validPredictionsAndLabels)
//通过train数据和test数据已经得到了一个原始predict,下面选择一个最优模型
val bestModel = cvModel.bestModel.asInstanceOf[PipelineModel]
//现在观察在train和交叉验证模型上的结果集
val results =
"=====================================================================================\r\n" +
s"Param trainSample: ${Preproessing.trainSample}\r\n" +
s"TrainData count : ${Preproessing.trainData.count}\r\n" +
s"ValidationData count : ${Preproessing.validationData.count}\r\n" +
s"TestData count : ${Preproessing.testData.count}\r\n" +
"\r\n===================================================================================\r\n" +
s"Param maxIter = ${MaxIter.mkString(",")}\r\n" +
s"Param numFolds = ${numFolds}\r\n" +
"\r\n===================================================================================\r\n" +
s"Train data MSE = ${trainRegressionMetrics.meanSquaredError}\r\n" +
s"Train data RMSE = ${trainRegressionMetrics.rootMeanSquaredError}\r\n" +
s"Train data R-squared = ${trainRegressionMetrics.r2}\r\n" +
s"Train data MEA = ${trainRegressionMetrics.meanAbsoluteError}\r\n" +
s"Train data Explained variance = ${trainRegressionMetrics.explainedVariance}\r\n" +
"\r\n===================================================================================\r\n" +
s"Validation data MSE = ${validRegressionMetrics.meanSquaredError}\r\n" +
s"Validation data RMSE = ${validRegressionMetrics.rootMeanSquaredError}\r\n" +
s"Validation data R-squared = ${validRegressionMetrics.r2}\r\n" +
s"Validation data MEA = ${validRegressionMetrics.meanAbsoluteError}\r\n" +
s"Validation data explained variance = ${validRegressionMetrics.explainedVariance}\r\n" +
"\r\n===================================================================================\r\n" +
s"CV params explained : ${cvModel.explainParams}n" +
s"LR params explained : ${bestModel.stages.last.asInstanceOf[LinearRegressionModel].explainParams}n" +
"\r\n==================================THE END==========================================="
println(results)
println("Run this prediction on test set")
cvModel.transform(Preproessing.testData)
.select("id", "prediction")
.withColumnRenamed("prediction", "loss")
.coalesce(1)
.write.format("com.databricks.spark.csv")
.save("file:\\C:\\Users\\PC\\Desktop\\墨菲斯文件备份\\Word文档\\学习资料\\spark\\书\\机器学习\\数据\\output\\res_LR.csv")
}
}
GBT算法代码
object ScalaGBT {
def main(args: Array[String]): Unit = {
//定义参数
val NumTrees = Seq(5, 10, 15)
val MaxBins = Seq(23)
val numFlods = 10
val MaxIters = Seq(10)
val MaxDepth = Seq(10)
val spark = SparkSessionCreate.createSession()
import spark.implicits._
val model: GBTRegressor = new GBTRegressor()
.setFeaturesCol("features")
.setLabelCol("label")
//build pipeline
val pipeline = new Pipeline().setStages((Preproessing.stringIndexerStages :+ Preproessing.assembler) :+ model)
//param builder
val paramGrid: Array[ParamMap] = new ParamGridBuilder()
.addGrid(model.maxIter, MaxIters)
.addGrid(model.maxBins, MaxBins)
.addGrid(model.maxDepth, MaxDepth)
.build()
println("Preparing K-fold Cross Vaildation and Grid Search")
//make CV
val cv: CrossValidator = new CrossValidator()
.setEstimator(pipeline)
.setEvaluator(new RegressionEvaluator)
.setEstimatorParamMaps(paramGrid)
println("Training model with GraidentBoostedTrees algorithm")
//train modeo with GBT
val cvModel = cv.fit(Preproessing.trainData)
println("Evaluating model on train and test data and calculating RMSE")
val trainPredictionsAndLabels: RDD[(Double, Double)] = cvModel.transform(Preproessing.trainData).select("label", "prediction").map { case Row(label: Double, prediction: Double) => (label, prediction) }.rdd
val validPredictionAndLabels: RDD[(Double, Double)] = cvModel.transform(Preproessing.validationData).select("label", "prediction").map { case Row(label: Double, prediction: Double) => (label, prediction) }.rdd
val trainRegressionMetrics = new RegressionMetrics(trainPredictionsAndLabels)
val validRegressionMetrics = new RegressionMetrics(validPredictionAndLabels)
//best model
val bestModel = cvModel.bestModel.asInstanceOf[PipelineModel]
val featureImportances = bestModel.stages.last.asInstanceOf[GBTRegressionModel].featureImportances.toArray
val FI_to_List_sorted = featureImportances.toList.sorted.toArray
val output =
"=====================================================================================\r\n" +
s"Param trainSample: ${Preproessing.trainSample}\r\n" +
s"Param testSample: ${Preproessing.testSample}\r\n" +
s"TrainData count : ${Preproessing.trainData.count}\r\n" +
s"ValidationData count : ${Preproessing.validationData.count}\r\n" +
s"TestData count : ${Preproessing.testData.count}\r\n" +
"\r\n===================================================================================\r\n" +
s"Param maxIter = ${MaxIters.mkString(",")}\r\n" +
s"Param numFolds = ${numFlods}\r\n" +
"\r\n===================================================================================\r\n" +
s"Train data MSE = ${trainRegressionMetrics.meanSquaredError}\r\n" +
s"Train data RMSE = ${trainRegressionMetrics.rootMeanSquaredError}\r\n" +
s"Train data R-squared = ${trainRegressionMetrics.r2}\r\n" +
s"Train data MEA = ${trainRegressionMetrics.meanAbsoluteError}\r\n" +
s"Train data Explained variance = ${trainRegressionMetrics.explainedVariance}\r\n" +
"\r\n===================================================================================\r\n" +
s"Validation data MSE = ${validRegressionMetrics.meanSquaredError}\r\n" +
s"Validation data RMSE = ${validRegressionMetrics.rootMeanSquaredError}\r\n" +
s"Validation data R-squared = ${validRegressionMetrics.r2}\r\n" +
s"Validation data MEA = ${validRegressionMetrics.meanAbsoluteError}\r\n" +
s"Validation data explained variance = ${validRegressionMetrics.explainedVariance}\r\n" +
"\r\n===================================================================================\r\n" +
s"CV params explained : ${cvModel.explainParams}n" +
s"GBT params explained : ${bestModel.stages.last.asInstanceOf[GBTRegressionModel].explainParams}n" +
s"GBT features importances : ${Preproessing.featureCols.zip(FI_to_List_sorted).map(x => s"x${x._1} = ${x._2}".mkString("n"))}\r\n" +
"\r\n==================================THE END==========================================="
println(output)
println("Run this prediction on test set")
cvModel.transform(Preproessing.testData)
.select("id", "prediction")
.withColumnRenamed("prediction", "loss")
.coalesce(1)
.write.format("com.databricks.spark.csv")
.save("file:\\C:\\Users\\PC\\Desktop\\墨菲斯文件备份\\Word文档\\学习资料\\spark\\书\\机器学习\\数据\\output\\res_GBT.csv")
}
}
RF代码
注意:RF代码中,如果运行时间过长,可以自行自定义设置一下MaxBins和MaxDepth和numfolds的参数,将其调小一点
object ScalaRF {
def main(args: Array[String]): Unit = {
//随机森林用于分类和回归
val spark = SparkSessionCreate.createSession()
import spark.implicits._
//定义参数
val NumTrees = Seq(5, 10, 15)
val MaxBins = Seq(23, 27, 30)
val numFolds = 10
val MaxIter: Seq[Int] = Seq(20)
val MaxDepth: Seq[Int] = Seq(20)
//建立 RF model
val model = new RandomForestRegressor().setFeaturesCol("features").setLabelCol("label")
//build pipeline
val pipeline: Pipeline = new Pipeline()
.setStages((Preproessing.stringIndexerStages :+ Preproessing.assembler) :+ model)
//build paramGrid
val paramGrid = new ParamGridBuilder()
.addGrid(model.numTrees, NumTrees)
.addGrid(model.maxDepth, MaxDepth)
.addGrid(model.maxBins, MaxBins)
.build()
println("Preparing K-fold Cross Validation and Grid Search : Model tuning")
val cv = new CrossValidator()
.setEstimator(pipeline)
.setEvaluator(new RegressionEvaluator)
.setNumFolds(numFolds)
.setEstimatorParamMaps(paramGrid)
//build cvmodel with RF training trainData
val cvModel = cv.fit(Preproessing.trainData)
println("Evaluating model on train and validation set and calculating RMSE")
val trainPredictionsAndLabels = cvModel.transform(Preproessing.trainData).select("label", "prediction").map { case Row(label: Double, prediction: Double) => (label, prediction) }.rdd
val validPredictionsAndLabels = cvModel.transform(Preproessing.validationData).select("label", "prediction").map { case Row(label: Double, prediction: Double) => (label, prediction) }.rdd
val trainRegressionMetrics = new RegressionMetrics(trainPredictionsAndLabels)
val validRegressionMetrics = new RegressionMetrics(validPredictionsAndLabels)
val bestModel = cvModel.bestModel.asInstanceOf[PipelineModel]
val featureImportances = bestModel.stages.last.asInstanceOf[RandomForestRegressionModel].featureImportances.toArray
val FI_to_List_sorted = featureImportances.toList.sorted.toArray
val output =
"=====================================================================================\r\n" +
s"Param trainSample: ${Preproessing.trainSample}\r\n" +
s"Param testSample: ${Preproessing.testSample}\r\n" +
s"TrainData count : ${Preproessing.trainData.count}\r\n" +
s"ValidationData count : ${Preproessing.validationData.count}\r\n" +
s"TestData count : ${Preproessing.testData.count}\r\n" +
"\r\n===================================================================================\r\n" +
s"Param maxIter = ${MaxIter.mkString(",")}\r\n" +
s"Param numFolds = ${numFolds}\r\n" +
"\r\n===================================================================================\r\n" +
s"Train data MSE = ${trainRegressionMetrics.meanSquaredError}\r\n" +
s"Train data RMSE = ${trainRegressionMetrics.rootMeanSquaredError}\r\n" +
s"Train data R-squared = ${trainRegressionMetrics.r2}\r\n" +
s"Train data MEA = ${trainRegressionMetrics.meanAbsoluteError}\r\n" +
s"Train data Explained variance = ${trainRegressionMetrics.explainedVariance}\r\n" +
"\r\n===================================================================================\r\n" +
s"Validation data MSE = ${validRegressionMetrics.meanSquaredError}\r\n" +
s"Validation data RMSE = ${validRegressionMetrics.rootMeanSquaredError}\r\n" +
s"Validation data R-squared = ${validRegressionMetrics.r2}\r\n" +
s"Validation data MEA = ${validRegressionMetrics.meanAbsoluteError}\r\n" +
s"Validation data explained variance = ${validRegressionMetrics.explainedVariance}\r\n" +
"\r\n===================================================================================\r\n" +
s"CV params explained : ${cvModel.explainParams}n" +
s"RF params explained : ${bestModel.stages.last.asInstanceOf[RandomForestRegressionModel].explainParams}n" +
s"RF features importances : ${Preproessing.featureCols.zip(FI_to_List_sorted).map(x => s"x${x._1} = ${x._2}".mkString("n"))}\r\n" +
"\r\n==================================THE END==========================================="
println(output)
println("Run this prediction on test set")
cvModel.transform(Preproessing.testData)
.select("id", "prediction")
.withColumnRenamed("prediction", "loss")
.coalesce(1)
.write.format("com.databricks.spark.csv")
.save("file:\\C:\\Users\\PC\\Desktop\\墨菲斯文件备份\\Word文档\\学习资料\\spark\\书\\机器学习\\数据\\output\\res_RF.csv")
}
}
输出结果
LR算法输出结果
=====================================================================================
Param trainSample: 1.0
TrainData count : 140977
ValidationData count : 47341
TestData count : 125546
===================================================================================
Param maxIter = 1000
Param numFolds = 10
===================================================================================
Train data MSE = 4523266.93398241
Train data RMSE = 2126.797342010378
Train data R-squared = -0.16181596223081596
Train data MEA = 1358.4888709703798
Train data Explained variance = 8415946.47720863
===================================================================================
Validation data MSE = 4651416.497204879
Validation data RMSE = 2156.714282700627
Validation data R-squared = -0.19498670604587942
Validation data MEA = 1358.6436775990019
Validation data explained variance = 8486835.155155173
===================================================================================
CV params explained : estimator: estimator for selection (current: pipeline_c5ad4ff638f1)
estimatorParamMaps: param maps for the estimator (current: [Lorg.apache.spark.ml.param.ParamMap;@17228435)
evaluator: evaluator used to select hyper-parameters that maximize the validated metric (current: regEval_1d803bd7fa7f)
numFolds: number of folds for cross validation (>= 2) (default: 3, current: 10)
seed: random seed (default: -1191137437)nLR params explained : aggregationDepth: suggested depth for treeAggregate (>= 2) (default: 2)
elasticNetParam: the ElasticNet mixing parameter, in range [0, 1]. For alpha = 0, the penalty is an L2 penalty. For alpha = 1, it is an L1 penalty (default: 0.0, current: 0.001)
featuresCol: features column name (default: features, current: features)
fitIntercept: whether to fit an intercept term (default: true)
labelCol: label column name (default: label, current: label)
maxIter: maximum number of iterations (>= 0) (default: 100, current: 1000)
predictionCol: prediction column name (default: prediction)
regParam: regularization parameter (>= 0) (default: 0.0, current: 0.001)
solver: the solver algorithm for optimization. If this is not set or empty, default value is 'auto' (default: auto)
standardization: whether to standardize the training features before fitting the model (default: true)
tol: the convergence tolerance for iterative algorithms (>= 0) (default: 1.0E-6, current: 1.0E-6)
weightCol: weight column name. If this is not set or empty, we treat all instance weights as 1.0 (undefined)n
==================================THE END===========================================
GBT算法输出结果
=====================================================================================
Param trainSample: 1.0
Param testSample: 1.0
TrainData count : 140977
ValidationData count : 47341
TestData count : 125546
===================================================================================
Param maxIter = 10
Param numFolds = 10
===================================================================================
Train data MSE = 2741423.427223714
Train data RMSE = 1655.7244418150365
Train data R-squared = 0.49488425396158653
Train data MEA = 1130.816386611616
Train data Explained variance = 8415946.477208637
===================================================================================
Validation data MSE = 5042438.8124634735
Validation data RMSE = 2245.5375330783213
Validation data R-squared = 0.12487009821777817
Validation data MEA = 1305.3855592766095
Validation data explained variance = 8487116.816812716
===================================================================================
CV params explained : estimator: estimator for selection (current: pipeline_4e0e35b462fe)
estimatorParamMaps: param maps for the estimator (current: [Lorg.apache.spark.ml.param.ParamMap;@6db173fc)
evaluator: evaluator used to select hyper-parameters that maximize the validated metric (current: regEval_eb8af9a6aee4)
numFolds: number of folds for cross validation (>= 2) (default: 3)
seed: random seed (default: -1191137437)nGBT params explained : cacheNodeIds: If false, the algorithm will pass trees to executors to match instances with nodes. If true, the algorithm will cache node IDs for each instance. Caching can speed up training of deeper trees. (default: false)
checkpointInterval: set checkpoint interval (>= 1) or disable checkpoint (-1). E.g. 10 means that the cache will get checkpointed every 10 iterations (default: 10)
featuresCol: features column name (default: features, current: features)
impurity: Criterion used for information gain calculation (case-insensitive). Supported options: variance (default: variance)
labelCol: label column name (default: label, current: label)
lossType: Loss function which GBT tries to minimize (case-insensitive). Supported options: squared, absolute (default: squared)
maxBins: Max number of bins for discretizing continuous features. Must be >=2 and >= number of categories for any categorical feature. (default: 32, current: 23)
maxDepth: Maximum depth of the tree. (>= 0) E.g., depth 0 means 1 leaf node; depth 1 means 1 internal node + 2 leaf nodes. (default: 5, current: 10)
maxIter: maximum number of iterations (>= 0) (default: 20, current: 10)
maxMemoryInMB: Maximum memory in MB allocated to histogram aggregation. (default: 256)
minInfoGain: Minimum information gain for a split to be considered at a tree node. (default: 0.0)
minInstancesPerNode: Minimum number of instances each child must have after split. If a split causes the left or right child to have fewer than minInstancesPerNode, the split will be discarded as invalid. Should be >= 1. (default: 1)
predictionCol: prediction column name (default: prediction)
seed: random seed (default: -131597770)
stepSize: Step size (a.k.a. learning rate) in interval (0, 1] for shrinking the contribution of each estimator. (default: 0.1)
subsamplingRate: Fraction of the training data used for learning each decision tree, in range (0, 1]. (default: 1.0)nGBT features importances : [Ljava.lang.String;@ee9832
==================================THE END===========================================
RF算法输出结果
这里我将部分参数调整:防止运行时间过长(文章后边会有这些参数的详细解释)
//定义参数
val NumTrees = Seq(5, 10, 15)
val MaxBins = Seq(23)
val numFolds = 5
val MaxIter: Seq[Int] = Seq(20)
val MaxDepth: Seq[Int] = Seq(10)
打印结果:
=====================================================================================
Param trainSample: 1.0
Param testSample: 1.0
TrainData count : 140977
ValidationData count : 47341
TestData count : 125546
===================================================================================
Param maxIter = 20
Param numFolds = 5
===================================================================================
Train data MSE = 3475665.49807464
Train data RMSE = 1864.3136801715102
Train data R-squared = 0.11756809943785507
Train data MEA = 1238.4531203300753
Train data Explained variance = 8415946.47746223
===================================================================================
Validation data MSE = 4296703.376504024
Validation data RMSE = 2072.8490964139246
Validation data R-squared = -0.13129110162667468
Validation data MEA = 1296.40526673937
Validation data explained variance = 8486770.991632275
===================================================================================
CV params explained : estimator: estimator for selection (current: pipeline_11f6cd05309a)
estimatorParamMaps: param maps for the estimator (current: [Lorg.apache.spark.ml.param.ParamMap;@7f50818d)
evaluator: evaluator used to select hyper-parameters that maximize the validated metric (current: regEval_6ebbc9328aa3)
numFolds: number of folds for cross validation (>= 2) (default: 3, current: 5)
seed: random seed (default: -1191137437)nRF params explained : cacheNodeIds: If false, the algorithm will pass trees to executors to match instances with nodes. If true, the algorithm will cache node IDs for each instance. Caching can speed up training of deeper trees. (default: false)
checkpointInterval: set checkpoint interval (>= 1) or disable checkpoint (-1). E.g. 10 means that the cache will get checkpointed every 10 iterations (default: 10)
featureSubsetStrategy: The number of features to consider for splits at each tree node. Supported options: auto, all, onethird, sqrt, log2, (0.0-1.0], [1-n]. (default: auto)
featuresCol: features column name (default: features, current: features)
impurity: Criterion used for information gain calculation (case-insensitive). Supported options: variance (default: variance)
labelCol: label column name (default: label, current: label)
maxBins: Max number of bins for discretizing continuous features. Must be >=2 and >= number of categories for any categorical feature. (default: 32, current: 23)
maxDepth: Maximum depth of the tree. (>= 0) E.g., depth 0 means 1 leaf node; depth 1 means 1 internal node + 2 leaf nodes. (default: 5, current: 10)
maxMemoryInMB: Maximum memory in MB allocated to histogram aggregation. (default: 256)
minInfoGain: Minimum information gain for a split to be considered at a tree node. (default: 0.0)
minInstancesPerNode: Minimum number of instances each child must have after split. If a split causes the left or right child to have fewer than minInstancesPerNode, the split will be discarded as invalid. Should be >= 1. (default: 1)
numTrees: Number of trees to train (>= 1) (default: 20, current: 15)
predictionCol: prediction column name (default: prediction)
seed: random seed (default: 235498149)
subsamplingRate: Fraction of the training data used for learning each decision tree, in range (0, 1]. (default: 1.0)nRF features importances : [Ljava.lang.String;@7811129f
==================================THE END===========================================
三种情况对比分析
参数对比
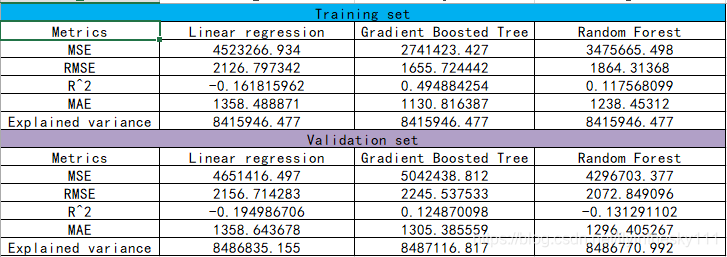
对比分析结论:
结论: 由上图可知,相比较LR和RF算法,GBT算法的精度更好,但是其实相比较LR而言,RF和GBT算法的精度都较它高 。(这里的RF算法由于受制于运行时间,参数和其余两者没有可比性,因此不具有太大的参考价值)。但是,从简单性来考虑,LR所耗资源和时间都比其余两者来的更少,更易于训练。而如果我们将RF算法和GBT算法的参数设置为一致的话,通过对比分析,可以发现,RF算法较GBT算法更优。(可以自行测试验证)。
结果输出比较
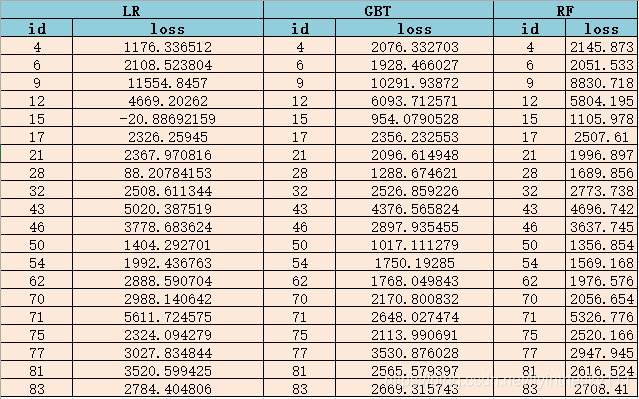
结语
对于一个小的训练数据集,LR模型更容易训练。然而,与GBT和随机森林模型相比,我们并没有获得更好的精度。因此,从表2中可以看出,我们不仅要预测保险理赔损失,还要预测保险理赔损失的产生,我们应该使用随机森林回归(RF)
下次文章内容简介:
后续将基于此次文章中代码部分的各个参数,以及整个机器学习的流程以及各个术语的解释进行详细的介绍。敬请期待!!!
如有问题欢迎添加作者微信:ljelzl416108 ,一同交流学习大数据和机器学习的知识!!!