#决策树集成
import mglearn
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
from sklearn.tree import DecisionTreeClassifier
from sklearn.datasets import load_breast_cancer
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
from sklearn.datasets import make_moons
from sklearn.ensemble import GradientBoostingClassifier
x,y = make_moons(n_samples=100,noise=0.25,random_state=3)
x_train,x_test,y_train,y_test = train_test_split(x,y,stratify=y,random_state=42)
forest = RandomForestClassifier(n_estimators=5,random_state=2)
forest.fit(x_train,y_train)
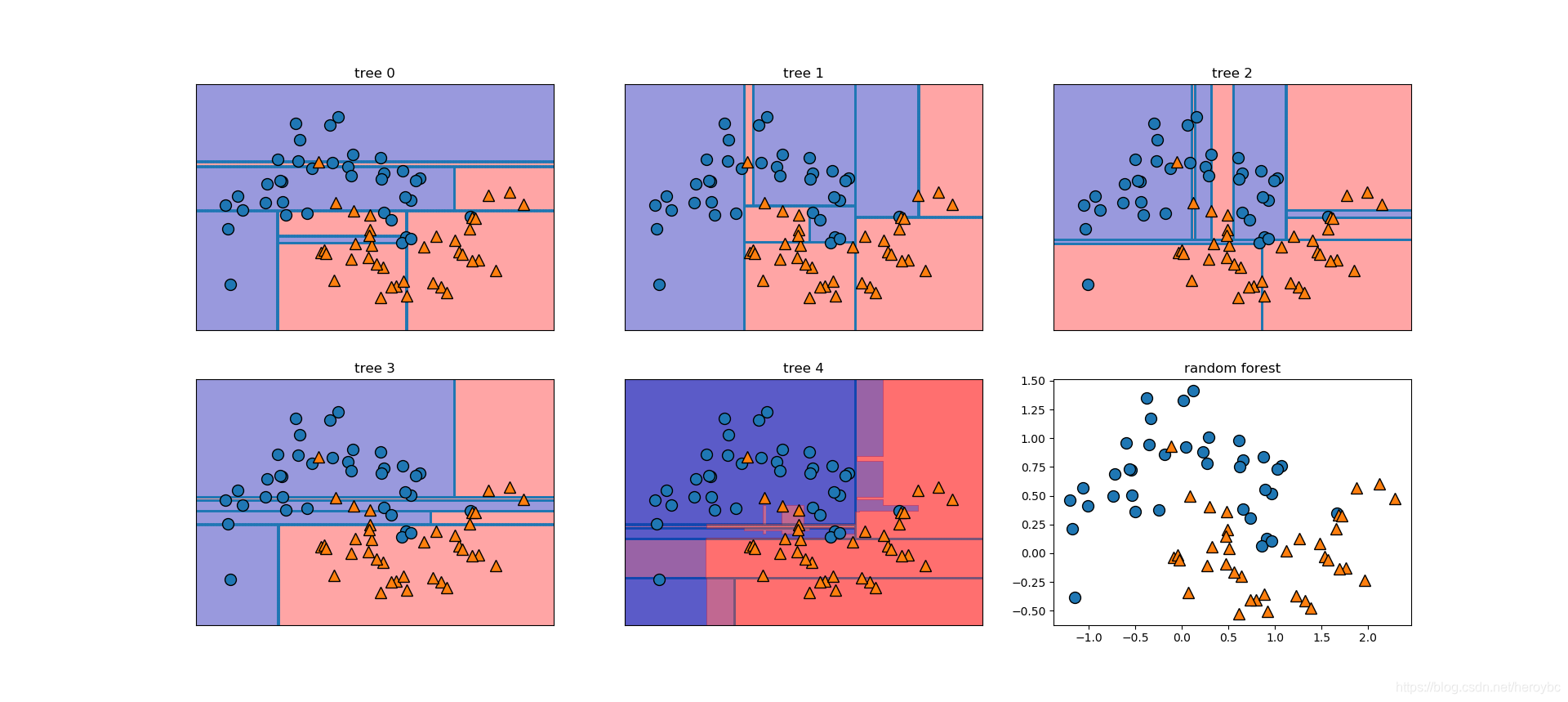
fig,axes = plt.subplots(2,3,figsize=(20,10))
for i,(ax,tree) in enumerate(zip(axes.ravel(),forest.estimators_)):
ax.set_title('tree {}'.format(i))
mglearn.plots.plot_tree_partition(x_train,y_train,tree,ax=ax)
mglearn.plots.plot_2d_separator(forest,x_train,fill=True,ax=axes[-1,1],alpha=.4)
axes[-1,-1].set_title('random forest')
mglearn.discrete_scatter(x_train[:,0],x_train[:,1],y_train)
cancer = load_breast_cancer()
x_train,x_test,y_train,y_test = train_test_split(cancer.data,cancer.target,random_state=0)
forest = RandomForestClassifier(n_estimators=100,random_state=0)
forest.fit(x_train,y_train)
print('accuracy on training set:{:.3f}'.format(forest.score(x_train,y_train)))
print('accuracy on test set:{:.3f}'.format(forest.score(x_test,y_test)))
def plot_feature_importances_cancer(model):
n_features = cancer.data.shape[1]
plt.barh(range(n_features),model.feature_importances_,align='center')
plt.yticks(np.arange(n_features),cancer.feature_names)
plt.xlabel('feature importance')
plt.ylabel("feature")
plot_feature_importances_cancer(forest)
accuracy on training set:1.000
accuracy on test set:0.972
def plot_feature_importances_cancer(model):
n_features = cancer.data.shape[1]
plt.barh(range(n_features),model.feature_importances_,align='center')
plt.yticks(np.arange(n_features),cancer.feature_names)
plt.xlabel('feature importance')
plt.ylabel("feature")
plot_feature_importances_cancer(forest)

from sklearn.ensemble import GradientBoostingClassifier
x_train,x_test,y_train,y_test = train_test_split(cancer.data,cancer.target,random_state=0)
gbrt = GradientBoostingClassifier(random_state=0)
gbrt.fit(x_train,y_train)
print('accuracy on training set:{:.3f}'.format(gbrt.score(x_train,y_train)))
print('accuracy on test set:{:.3f}'.format(gbrt.score(x_test,y_test)))
accuracy on training set:1.000
accuracy on test set:0.958
gbrt = GradientBoostingClassifier(random_state=0,max_depth=1)
gbrt.fit(x_train,y_train)
print('accuracy on training set:{:.3f}'.format(gbrt.score(x_train,y_train)))
print('accuracy on test set:{:.3f}'.format(gbrt.score(x_test,y_test)))
plot_feature_importances_cancer(gbrt)
accuracy on training set:0.991
accuracy on test set:0.972

gbrt = GradientBoostingClassifier(random_state=0,learning_rate=0.01)
gbrt.fit(x_train,y_train)
print('accuracy on training set:{:.3f}'.format(gbrt.score(x_train,y_train)))
print('accuracy on test set:{:.3f}'.format(gbrt.score(x_test,y_test)))
accuracy on training set:0.988
accuracy on test set:0.965
全文代码:
#决策树集成
import mglearn
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
from sklearn.tree import DecisionTreeClassifier
from sklearn.datasets import load_breast_cancer
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
from sklearn.datasets import make_moons
x,y = make_moons(n_samples=100,noise=0.25,random_state=3)
x_train,x_test,y_train,y_test = train_test_split(x,y,stratify=y,random_state=42)
forest = RandomForestClassifier(n_estimators=5,random_state=2)
forest.fit(x_train,y_train)
fig,axes = plt.subplots(2,3,figsize=(20,10))
for i,(ax,tree) in enumerate(zip(axes.ravel(),forest.estimators_)):
ax.set_title('tree {}'.format(i))
mglearn.plots.plot_tree_partition(x_train,y_train,tree,ax=ax)
mglearn.plots.plot_2d_separator(forest,x_train,fill=True,ax=axes[-1,1],alpha=.4)
axes[-1,-1].set_title('random forest')
mglearn.discrete_scatter(x_train[:,0],x_train[:,1],y_train)
cancer = load_breast_cancer()
x_train,x_test,y_train,y_test = train_test_split(cancer.data,cancer.target,random_state=0)
forest = RandomForestClassifier(n_estimators=100,random_state=0)
forest.fit(x_train,y_train)
print('accuracy on training set:{:.3f}'.format(forest.score(x_train,y_train)))
print('accuracy on test set:{:.3f}'.format(forest.score(x_test,y_test)))
def plot_feature_importances_cancer(model):
n_features = cancer.data.shape[1]
plt.barh(range(n_features),model.feature_importances_,align='center')
plt.yticks(np.arange(n_features),cancer.feature_names)
plt.xlabel('feature importance')
plt.ylabel("feature")
plot_feature_importances_cancer(forest)
from sklearn.ensemble import GradientBoostingClassifier
x_train,x_test,y_train,y_test = train_test_split(cancer.data,cancer.target,random_state=0)
gbrt = GradientBoostingClassifier(random_state=0)
gbrt.fit(x_train,y_train)
print('accuracy on training set:{:.3f}'.format(gbrt.score(x_train,y_train)))
print('accuracy on test set:{:.3f}'.format(gbrt.score(x_test,y_test)))
gbrt = GradientBoostingClassifier(random_state=0,max_depth=1)
gbrt.fit(x_train,y_train)
print('accuracy on training set:{:.3f}'.format(gbrt.score(x_train,y_train)))
print('accuracy on test set:{:.3f}'.format(gbrt.score(x_test,y_test)))
plot_feature_importances_cancer(gbrt)
gbrt = GradientBoostingClassifier(random_state=0,learning_rate=0.01)
gbrt.fit(x_train,y_train)
print('accuracy on training set:{:.3f}'.format(gbrt.score(x_train,y_train)))
print('accuracy on test set:{:.3f}'.format(gbrt.score(x_test,y_test)))
