Several new pre-trained contextualized embeddings are released in 2018. New state-of-the-art results is changing every month. BERT is one of the famous model. In this story, we will extend BERT to see how we can apply BERT on different domain problem.
Allen Institute for Artificial Intelligence (AI2) further study on BERT and released SciBERT which is based on BERT to address the performance on scientific data. It uses a pre-trained model from BERT and fine-tune contextualized embeddings by using scientific publications which including 18% papers from computer science domain and 82% from the broad biomedical domain. On the other hand, Lee et al. work on biomedical domain. They also noticed that generic pretrained NLP model may not work very well in specific domain data. Therefore, they fine-tuned BERT to be BioBERT and 0.51% ~ 9.61% absolute improvement in biomedical’s NER, relation extraction and question answering NLP tasks.
This story will discuss about SCIBERT: Pretrained Contextualized Embeddings for Scientific Text (Beltagy et al., 2019), BioBERT: a pre-trained biomedical language representation model for biomedical text mining (Lee et al., 2019). The following are will be covered:
- Data
- Architecture
- Experiment
Data
Both SciBERT and BioBERT also introduce domain specific data for pre-training. Beltag et al. use 1.14M papers are random pick from Semantic Scholar to fine-tune BERT and building SciBERT. The corpus includes 18% computer science domain paper and 82% broad biomedical domain papers. On the other hand, Lee et al. use BERT’s original training data which includes English Wikipedia and BooksCorpus and domain specific data which are PubMed abstracts and PMC full text articles to fine-tuning BioBERT model.
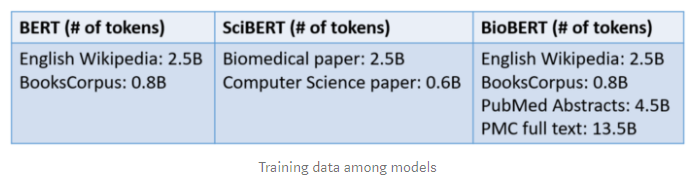
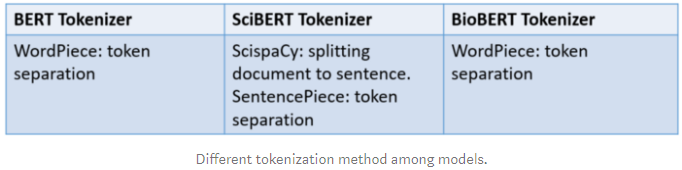
Some changes are applied to make a successful in scientific text. ScispaCy, ascientific specific version of spaCy, is leveraged to split document to sentences. After that Beltagy et al. use SentencePiece library to build new WordPiece vocabulary for SciBERT rather than using BERT’s vocabulary.
Architecture
Both SciBERT and BioBERT follow BERT model architecture which is multi bidirectional transformer and learning text representation by predicting masked token and next sentence. A sequence of tokens will be transform to token embeddings, segment embeddings and position embeddings. Token embeddings refers to contextualized word embeddings, segment embeddings only include 2 embeddings which are either 0 or 1 to represent first sentence and second sentence, position embeddings stores the token position relative to the sequence. You can visit this story to understand more about BERT.

Experiment
Scientific BERT (SciBERT)
Both Named Entity Recognition (NER) and Participant Intervention Comparison Outcome Extraction (PICO) are sequence labelling. Dependency Parsing (DEP) is predicting the dependencies between tokens in the sentence. Classification (CLS) and Relation Classification (REL) are classification tasks.



Biomedical BERT (BioBERT)
From below table, you can noticed that BioBERT outperform BERT on domain specific dataset.


Take Away
- Universal pretrained model may not able to achieve the state-of-the-art result in specific domain. Therefore, fine-tuned step is necessary to boost up performance on target dataset.
- Transformer (multiple self attentions) become more and more famous after BERT and BERT’s based model. Also noticed that BERT’s based model keep achieve state-of-the-art performance.
About Me
I am Data Scientist in Bay Area. Focusing on state-of-the-art in Data Science, Artificial Intelligence , especially in NLP and platform related. Feel free to connect with me on LinkedIn or following me on Medium or Github.
Extension Reading
Reference
- I. Beltagy, A. Cohan and K. Lo. SCIBERT: Pretrained Contextualized Embeddings for Scientific Text. 2019
- J. Lee, W. Yoon, S. Kim, D. Kim, S. Kim, C. H. So and J. Kang. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. 2019
Refer: https://towardsdatascience.com/how-to-apply-bert-in-scientific-domain-2d9db0480bd9
