版权声明:本文为博主原创文章,未经博主允许不得转载。 https://blog.csdn.net/wangyaninglm/article/details/87540653
未完待续
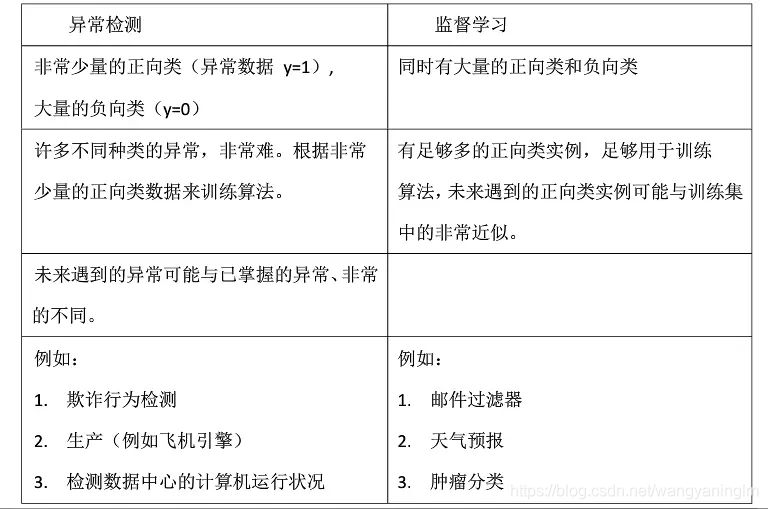
什么是异常检测
https://blog.csdn.net/App_12062011/article/details/84797641
数据加载
Meanshift 聚类
从源代码可以看出:
https://github.com/scikit-learn/scikit-learn/blob/7389dba/sklearn/cluster/mean_shift_.py#L298
<class ‘sklearn.cluster.mean_shift_.MeanShift’>
中显示不能分类的数据其类别标签就是-1
def mean_shift(X, bandwidth=None, seeds=None, bin_seeding=False,
min_bin_freq=1, cluster_all=True, max_iter=300,
n_jobs=None):
"""Perform mean shift clustering of data using a flat kernel.
Read more in the :ref:`User Guide <mean_shift>`.
Parameters
----------
X : array-like, shape=[n_samples, n_features]
Input data.
bandwidth : float, optional
Kernel bandwidth.
If bandwidth is not given, it is determined using a heuristic based on
the median of all pairwise distances. This will take quadratic time in
the number of samples. The sklearn.cluster.estimate_bandwidth function
can be used to do this more efficiently.
seeds : array-like, shape=[n_seeds, n_features] or None
Point used as initial kernel locations. If None and bin_seeding=False,
each data point is used as a seed. If None and bin_seeding=True,
see bin_seeding.
bin_seeding : boolean, default=False
If true, initial kernel locations are not locations of all
points, but rather the location of the discretized version of
points, where points are binned onto a grid whose coarseness
corresponds to the bandwidth. Setting this option to True will speed
up the algorithm because fewer seeds will be initialized.
Ignored if seeds argument is not None.
min_bin_freq : int, default=1
To speed up the algorithm, accept only those bins with at least
min_bin_freq points as seeds.
cluster_all : boolean, default True
If true, then all points are clustered, even those orphans that are
not within any kernel. Orphans are assigned to the nearest kernel.
If false, then orphans are given cluster label -1.
max_iter : int, default 300
Maximum number of iterations, per seed point before the clustering
operation terminates (for that seed point), if has not converged yet.
n_jobs : int or None, optional (default=None)
The number of jobs to use for the computation. This works by computing
each of the n_init runs in parallel.
``None`` means 1 unless in a :obj:`joblib.parallel_backend` context.
``-1`` means using all processors. See :term:`Glossary <n_jobs>`
for more details.
.. versionadded:: 0.17
Parallel Execution using *n_jobs*.
Returns
-------
cluster_centers : array, shape=[n_clusters, n_features]
Coordinates of cluster centers.
labels : array, shape=[n_samples]
Cluster labels for each point.
Notes
-----
For an example, see :ref:`examples/cluster/plot_mean_shift.py
<sphx_glr_auto_examples_cluster_plot_mean_shift.py>`.
"""
if bandwidth is None:
bandwidth = estimate_bandwidth(X, n_jobs=n_jobs)
elif bandwidth <= 0:
raise ValueError("bandwidth needs to be greater than zero or None,\
got %f" % bandwidth)
if seeds is None:
if bin_seeding:
seeds = get_bin_seeds(X, bandwidth, min_bin_freq)
else:
seeds = X
n_samples, n_features = X.shape
center_intensity_dict = {}
# We use n_jobs=1 because this will be used in nested calls under
# parallel calls to _mean_shift_single_seed so there is no need for
# for further parallelism.
nbrs = NearestNeighbors(radius=bandwidth, n_jobs=1).fit(X)
# execute iterations on all seeds in parallel
all_res = Parallel(n_jobs=n_jobs)(
delayed(_mean_shift_single_seed)
(seed, X, nbrs, max_iter) for seed in seeds)
# copy results in a dictionary
for i in range(len(seeds)):
if all_res[i] is not None:
center_intensity_dict[all_res[i][0]] = all_res[i][1]
if not center_intensity_dict:
# nothing near seeds
raise ValueError("No point was within bandwidth=%f of any seed."
" Try a different seeding strategy \
or increase the bandwidth."
% bandwidth)
# POST PROCESSING: remove near duplicate points
# If the distance between two kernels is less than the bandwidth,
# then we have to remove one because it is a duplicate. Remove the
# one with fewer points.
sorted_by_intensity = sorted(center_intensity_dict.items(),
key=lambda tup: (tup[1], tup[0]),
reverse=True)
sorted_centers = np.array([tup[0] for tup in sorted_by_intensity])
unique = np.ones(len(sorted_centers), dtype=np.bool)
nbrs = NearestNeighbors(radius=bandwidth,
n_jobs=n_jobs).fit(sorted_centers)
for i, center in enumerate(sorted_centers):
if unique[i]:
neighbor_idxs = nbrs.radius_neighbors([center],
return_distance=False)[0]
unique[neighbor_idxs] = 0
unique[i] = 1 # leave the current point as unique
cluster_centers = sorted_centers[unique]
# ASSIGN LABELS: a point belongs to the cluster that it is closest to
nbrs = NearestNeighbors(n_neighbors=1, n_jobs=n_jobs).fit(cluster_centers)
labels = np.zeros(n_samples, dtype=np.int)
distances, idxs = nbrs.kneighbors(X)
if cluster_all:
labels = idxs.flatten()
else:
labels.fill(-1)
bool_selector = distances.flatten() <= bandwidth
labels[bool_selector] = idxs.flatten()[bool_selector]
return cluster_centers, labels
代码注释参考:
https://blog.csdn.net/jiaqiangbandongg/article/details/53557500