介绍:
GitHub - EBI-Metagenomics/EukCC: Tool to estimate genome quality of microbial eukaryotes
安装:
docker:
docker pull microbiomeinformatics/eukcc推荐conda 环境:
conda install -c conda-forge -c bioconda "eukcc>=2"
# mamba更快
mamba install -c conda-forge -c bioconda "eukcc>=2"
pip install eukcc数据库配置,docker记得映射目录
mkdir eukccdb
cd eukccdb
wget http://ftp.ebi.ac.uk/pub/databases/metagenomics/eukcc/eukcc2_db_ver_1.1.tar.gz
tar -xzvf eukcc2_db_ver_1.1.tar.gz数据库下载地址:Index of /pub/databases/metagenomics/eukcc
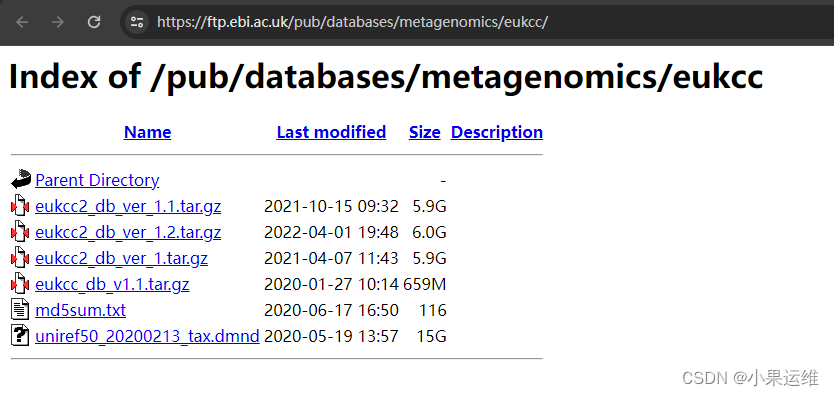
下载数据库注意版本,一般选版本2吧,
链接:https://ftp.ebi.ac.uk/pub/databases/metagenomics/eukcc/eukcc2_db_ver_1.2.tar.gz
https://ftp.ebi.ac.uk/pub/databases/metagenomics/eukcc/eukcc_db_v1.1.tar.gz
还有个副产品,diamond的数据库,不过好像看不出是diamond的哪个版本生成的,用的时候不好用的话就用下载的数据库再生成一遍吧。
https://ftp.ebi.ac.uk/pub/databases/metagenomics/eukcc/uniref50_20200213_tax.dmnd
如果不知道数据库位置,或者软件找不到位置,那就简单吧,设置DB目录
export EUKCC2_DB=/path/to/.../eukcc2_db_ver_1.1快速开始
#EukCC on a single MAG
#We assume that you did set you $EUKCC2_DB to the correct location. If not please use the --db flag to pass the database to EukCC.
eukcc single --out outfolder --threads 8 bin.fa
#EukCC will then run on 8 threads. You can pass nucleotide fastas or proteomes to EukCC. It will automatically try to detect if it has to predict proteins or not.
#By default it will never use more than a single threads for placing the genomes in the reference tree, to save memory.
#EukCC on a folder of bins
eukcc folder --out outfolder --threads 8 bins
#EukCC will assume that the folder contains files with the suffix .fa. If that is not the case please adjust the parameter.序列拼接流程
双端序列需要先构建bam索引
cat binfolder/*.fa > pseudo_contigs.fasta
bwa index pseudo_contigs.fasta
bwa mem -t 8 pseudo_contigs.fasta reads_1.fastq.gz reads_2.fastq.gz |
samtools view -q 20 -Sb - |
samtools sort -@ 8 -O bam - -o alignment.bam
samtools index alignment.bam利用py脚本生成关联表
binlinks.py --ANI 99 --within 1500 \
--out linktable.csv binfolder alignment.bamIf you have multiple bam files, pass all of them to the script (e.g. *.bam).
You will obtain a three column file (bin_1,bin_2,links).
拼接bins
eukcc folder \
--out outfolder \
--threads 8 \
--links linktable.csv \
binfolderEukCC 首先将分别对所有bins进行运行。随后,它会识别那些至少达到50%完整度但尚未超过100-improve_percent的中等质量bins。接下来,它会找出那些通过至少100对端读配对与这些中等质量bins相连接的bins。若经过合并后bin的质量评分有所提高,则该bin将会被合并。
已合并的bins可以在输出文件夹中找到。
警示
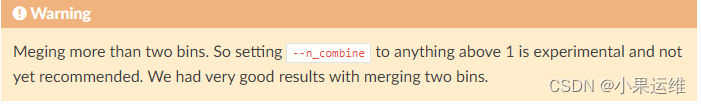
blinks.py
#!/usr/bin/env python3
import pysam
from Bio import SeqIO
from collections import defaultdict
import os
import argparse
import logging
import csv
def is_in(read, contig_map, within=1000):
if read.reference_name not in contig_map.keys():
return False
if read.reference_start <= within or read.reference_end <= within:
return True
elif read.reference_start > (
contig_map[read.reference_name] - within
) or read.reference_end > (contig_map[read.reference_name] - within):
return True
else:
return False
def keep_read(read, contig_map, within=1000, min_ANI=98, min_cov=0):
ani = (
(read.query_alignment_length - read.get_tag("NM"))
/ float(read.query_alignment_length)
* 100
)
cov = read.query_alignment_length / float(read.query_length) * 100
if ani >= min_ANI and cov >= min_cov and is_in(read, contig_map, within) is True:
return True
else:
return False
def contig_map(bindir, suffix=".fa"):
m = {}
for f in os.listdir(bindir):
if f.endswith(suffix) is False:
continue
path = os.path.join(bindir, f)
with open(path, "r") as handle:
for record in SeqIO.parse(handle, "fasta"):
m[record.name] = len(record.seq)
return m
def bin_map(bindir, suffix=".fa"):
contigs = defaultdict(str)
contigs_per_bin = defaultdict(int)
for f in os.listdir(bindir):
if f.endswith(suffix) is False:
continue
path = os.path.join(bindir, f)
binname = os.path.basename(f)
with open(path, "r") as handle:
for record in SeqIO.parse(handle, "fasta"):
contigs[record.name] = binname
contigs_per_bin[binname] += 1
return contigs, contigs_per_bin
def read_pair_generator(bam):
"""
Generate read pairs in a BAM file or within a region string.
Reads are added to read_dict until a pair is found.
From: https://www.biostars.org/p/306041/
"""
read_dict = defaultdict(lambda: [None, None])
for read in bam.fetch():
if not read.is_paired or read.is_secondary or read.is_supplementary:
continue
qname = read.query_name
if qname not in read_dict:
if read.is_read1:
read_dict[qname][0] = read
else:
read_dict[qname][1] = read
else:
if read.is_read1:
yield read, read_dict[qname][1]
else:
yield read_dict[qname][0], read
del read_dict[qname]
def read_bam_file(bamf, link_table, cm, within, ANI):
samfile = pysam.AlignmentFile(bamf, "rb")
# generate link table
logging.info("Parsing Bam file. This can take a few moments")
for read, mate in read_pair_generator(samfile):
if keep_read(read, cm, within, min_ANI=ANI) and keep_read(
mate, cm, within, min_ANI=ANI
):
# fill in the table
link_table[read.reference_name][mate.reference_name] += 1
if read.reference_name != mate.reference_name:
link_table[mate.reference_name][read.reference_name] += 1
return link_table
def main():
# set arguments
# arguments are passed to classes
parser = argparse.ArgumentParser(
description="Evaluate completeness and contamination of a MAG."
)
parser.add_argument("bindir", type=str, help="Run script on these bins")
parser.add_argument(
"bam",
type=str,
help="Bam file(s) with reads aligned against all contigs making up the bins",
nargs="+",
)
parser.add_argument(
"--out",
"-o",
type=str,
required=False,
help="Path to output table (Default: links.csv)",
default="links.csv",
)
parser.add_argument(
"--ANI", type=float, required=False, help="ANI of matching read", default=99
)
parser.add_argument(
"--within",
type=int,
required=False,
help="Within this many bp we need the read to map",
default=1000,
)
parser.add_argument(
"--contigs",
"-c",
action="store_true",
default=False,
help="Instead of bins print contigs",
)
parser.add_argument(
"--quiet",
"-q",
dest="quiet",
action="store_true",
default=False,
help="Silcence most output",
)
parser.add_argument(
"--debug",
"-d",
action="store_true",
default=False,
help="Debug and thus ignore safety",
)
args = parser.parse_args()
# define logging
logLevel = logging.INFO
if args.quiet:
logLevel = logging.WARNING
elif args.debug:
logLevel = logging.DEBUG
logging.basicConfig(
format="%(asctime)s %(message)s",
datefmt="%d-%m-%Y %H:%M:%S: ",
level=logLevel,
)
bindir = args.bindir
cm = contig_map(bindir)
bm, contigs_per_bin = bin_map(bindir)
logging.debug("Found {} contigs".format(len(cm)))
link_table = defaultdict(lambda: defaultdict(int))
bin_table = defaultdict(lambda: defaultdict(int))
# iterate all bam files
for bamf in args.bam:
link_table = read_bam_file(bamf, link_table, cm, args.within, args.ANI)
logging.debug("Created link table with {} entries".format(len(link_table)))
# generate bin table
for contig_1, dic in link_table.items():
for contig_2, links in dic.items():
bin_table[bm[contig_1]][bm[contig_2]] += links
logging.debug("Created bin table with {} entries".format(len(bin_table)))
out_data = []
logging.debug("Constructing output dict")
if args.contigs:
for contig_1, linked in link_table.items():
for contig_2, links in linked.items():
out_data.append(
{
"bin_1": bm[contig_1],
"bin_2": bm[contig_2],
"contig_1": contig_1,
"contig_2": contig_2,
"links": links,
"bin_1_contigs": contigs_per_bin[bm[contig_1]],
"bin_2_contigs": contigs_per_bin[bm[contig_2]],
}
)
else:
for bin_1, dic in bin_table.items():
for bin_2, links in dic.items():
out_data.append({"bin_1": bin_1, "bin_2": bin_2, "links": links})
logging.debug("Out data has {} rows".format(len(out_data)))
# results
logging.info("Writing output")
with open(args.out, "w") as fout:
if len(out_data) > 0:
cout = csv.DictWriter(fout, fieldnames=list(out_data[0].keys()))
cout.writeheader()
for row in out_data:
cout.writerow(row)
else:
logging.warning("No rows to write")
if __name__ == "__main__":
main()scripts/filter_euk_bins.py
#!/usr/bin/env python3
#
# This file is part of the EukCC (https://github.com/openpaul/eukcc).
# Copyright (c) 2019 Paul Saary
#
# This program is free software: you can redistribute it and/or modify
# it under the terms of the GNU General Public License as published by
# the Free Software Foundation, version 3.
#
# This program is distributed in the hope that it will be useful, but
# WITHOUT ANY WARRANTY; without even the implied warranty of
# MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU
# General Public License for more details.
#
# You should have received a copy of the GNU General Public License
# along with this program. If not, see <http://www.gnu.org/licenses/>.
# provides all file operation functions
# used inthis package
import os
import argparse
import subprocess
import logging
import tempfile
import gzip
from multiprocessing import Pool
# backup fasta handler, so we can use readonly directories
class fa_class:
def __init__(self, seq, name, long_name):
self.seq = seq
self.name = name
self.long_name = long_name
def __str__(self):
return self.seq
def __len__(self):
return len(self.seq)
def Fasta(path):
"""
Iterator for fasta files
"""
entry = False
with open(path) as fin:
for line in fin:
if line.startswith(">"):
if entry is not False:
entry.seq = "".join(entry.seq)
yield entry
# define new entry
long_name = line.strip()[1:]
name = long_name.split()[0]
entry = fa_class([], name, long_name)
else:
entry.seq.append(line.strip())
# yield last one
entry.seq = "".join(entry.seq)
yield entry
def gunzip(path, tmp_dir):
"""
Gunzip a file for EukRep
"""
if path.endswith(".gz"):
fna_path = os.path.join(tmp_dir, "contigs.fna")
logging.debug("Going to unzip fasta into {}".format(fna_path))
with gzip.open(path, "r") as fin, open(fna_path, "w") as fout:
for line in fin:
fout.write(line.decode())
path = fna_path
logging.debug("Done unzipping {}".format(fna_path))
return path
class EukRep:
"""Class to call and handle EukRep data"""
def __init__(self, fasta, eukout, bacout=None, minl=1500, tie="euk"):
self.fasta = fasta
self.eukout = eukout
self.bacout = bacout
self.minl = minl
self.tie = tie
def run(self):
# command list will be called
cmd = [
"EukRep",
"--min",
str(self.minl),
"-i",
self.fasta,
"--seq_names",
"-ff",
"--tie",
self.tie,
"-o",
self.eukout,
]
if self.bacout is not None:
cmd.extend(["--prokarya", self.bacout])
subprocess.run(cmd, check=True, shell=False)
self.read_result()
def read_result(self):
self.euks = self.read_eukfile(self.eukout)
self.bacs = set()
if self.bacout is not None:
self.bacs = self.read_eukfile(self.bacout)
def read_eukfile(self, path):
lst = set()
with open(path) as infile:
for line in infile:
lst.add(line.strip())
return lst
class bin:
def __init__(self, path, eukrep):
self.e = eukrep
self.bname = os.path.basename(path)
self.path = os.path.abspath(path)
def __str__(self):
return "{} euks: {} bacs: {}".format(self.bname, self.table["euks"], self.table["bacs"])
def stats(self):
"""read bin content and figure genomic composition"""
logging.debug("Loading bin")
fa_file = Fasta(self.path)
stats = {"euks": 0, "bacs": 0, "NA": 0, "sum": 0}
# loop and compute stats
logging.debug("Make per bin stats")
for seq in fa_file:
if seq.name in self.e.euks:
stats["euks"] += len(seq)
elif seq.name in self.e.bacs:
stats["bacs"] += len(seq)
else:
stats["NA"] += len(seq)
stats["sum"] = sum([v for k, v in stats.items()])
self.table = stats
def decide(self, eukratio=0.2, bacratio=0.1, minbp=100000, minbpeuks=1000000):
"""
rule to handle decision logic
"""
keep = True
allb = self.table["sum"]
if self.table["euks"] < minbpeuks:
keep = False
logging.info(f"Eukaryotic DNA amount only {self.table['euks']} instead of target {minbpeuks}")
elif self.table["euks"] / allb <= eukratio:
keep = False
logging.info(f"Eukaryotic DNA ratio not higher than {eukratio}")
elif self.table["bacs"] / allb >= bacratio:
keep = False
logging.info(f"Bacterial DNA content higher than {bacratio}")
elif self.table["sum"] < minbp:
keep = False
logging.info("We did not find at least %d bp of DNA", minbp)
self.keep = keep
if __name__ == "__main__":
parser = argparse.ArgumentParser()
parser.add_argument("--output", help="path for the output table", default="assignment.csv", type=str)
parser.add_argument("bins", nargs="+", help="all bins to classify", type=str)
parser.add_argument(
"--threads",
"-t",
type=int,
help="How many bins should be run in parallel (Default: 1)",
default=1,
)
parser.add_argument(
"--minl",
type=int,
help="define minimal length of contig for EukRep \
to classify (default: 1500)",
default=1500,
)
parser.add_argument(
"--eukratio",
type=float,
help="This ratio of eukaryotic DNA to all DNA has to be found\
at least (default: 0, ignore)",
default=0,
)
parser.add_argument(
"--bacratio",
type=float,
help="discard bins with bacterial ratio of higher than\
(default: 1, ignore)",
default=1,
)
parser.add_argument(
"--minbp",
type=float,
help="Only keep bins with at least n bp of dna\
(default: 8000000)",
default=8000000,
)
parser.add_argument(
"--minbpeuks",
type=float,
help="Only keep bins with at least n bp of Eukaryotic dna\
(default: 5000000)",
default=5000000,
)
parser.add_argument("--rerun", action="store_true", help="rerun even if output exists", default=False)
parser.add_argument("--quiet", action="store_true", help="supress information", default=False)
parser.add_argument("--debug", action="store_true", help="Make it more verbose", default=False)
args = parser.parse_args()
# define logging
logLevel = logging.INFO
if args.quiet:
logLevel = logging.WARNING
elif args.debug:
logLevel = logging.DEBUG
logging.basicConfig(format="%(asctime)s %(message)s", datefmt="%m/%d/%Y %H:%M:%S: ", level=logLevel)
def evaluate_bin(path):
if not os.path.exists(path):
logging.error("Can not find file {}".format(path))
exit(1)
logging.info("Launch on {}".format(path))
with tempfile.TemporaryDirectory(prefix="filter_EukRep_") as tmp_dir:
logging.debug("Using tmp folder: {}".format(tmp_dir))
eukfile = os.path.join(tmp_dir, "euks.fna")
bacfile = os.path.join(tmp_dir, "bacs.fna")
# EukRep can not deal with Gzipped Fasta files, so we unzip it in case it is a Gzip file
path = gunzip(path, tmp_dir)
# Launching EukRep
logging.debug(f"Starting EukRep on {path}")
eukrep_result = EukRep(path, eukfile, bacfile, minl=args.minl)
eukrep_result.run()
b = bin(path, eukrep_result)
b.stats()
b.decide(eukratio=args.eukratio, bacratio=args.bacratio, minbp=args.minbp, minbpeuks=args.minbpeuks)
return b
# multithreading pool
pool = Pool(processes=args.threads)
results = pool.map(evaluate_bin, args.bins)
pool.close()
pool.join()
with open(args.output, "w") as outfile:
outfile.write("path,binname,passed,bp_eukaryote,bp_prokaryote,bp_unassigned,bp_sum\n")
for b in results:
outfile.write(
f"{b.path},{b.bname},{b.keep},{b.table['euks']},{b.table['bacs']},{b.table['NA']},{b.table['sum']}\n"
)