分类与回归
文章目录
一、分类
1. 逻辑分类
Logistic回归是一种用于预测分类响应的流行方法。 这是广义线性模型的一种特殊情况,可以预测结果的可能性。 在spark.ml中,逻辑回归可以通过使用二项式逻辑回归来预测二进制结果,或者可以通过使用多项逻辑回归来预测多类结果。 使用family参数在这两种算法之间进行选择,或者不设置它,Spark会推断出正确的变体。
通过将family参数设置为“multinomial”,可以将多项式逻辑回归用于二进制分类。 它将产生两组系数和两个截距。
当对具有恒定非零列的数据集进行LogisticRegressionModel拟合而没有截距时,Spark MLlib为恒定非零列输出零系数。 此行为与R glmnet相同,但与LIBSVM不同。
1.1 二元逻辑回归
有关二项式逻辑回归的实现的更多背景和更多详细信息,请参阅spark.mllib中的逻辑回归文档。
from pyspark.ml.classification import LogisticRegression
# Load training data
training = spark.read.format("libsvm").load("data/mllib/sample_libsvm_data.txt")
lr = LogisticRegression(maxIter=10, regParam=0.3, elasticNetParam=0.8)
# Fit the model
lrModel = lr.fit(training)
# Print the coefficients and intercept for logistic regression
print("Coefficients: " + str(lrModel.coefficients))
print("Intercept: " + str(lrModel.intercept))
# We can also use the multinomial family for binary classification
mlr = LogisticRegression(maxIter=10, regParam=0.3, elasticNetParam=0.8, family="multinomial")
# Fit the model
mlrModel = mlr.fit(training)
# Print the coefficients and intercepts for logistic regression with multinomial family
print("Multinomial coefficients: " + str(mlrModel.coefficientMatrix))
print("Multinomial intercepts: " + str(mlrModel.interceptVector))
LogisticRegressionTrainingSummary提供LogisticRdgressionModel的总结。在二进制分类的情况下,某些其他指标可用,例如ROC曲线。可以通过binarySummary方法访问binary summary
from pyspark.ml.classification import LogisticRegression
# Extract the summary from the returned LogisticRegressionModel instance trained
# in the earlier example
trainingSummary = lrModel.summary
# Obtain the objective per iteration
objectiveHistory = trainingSummary.objectiveHistory
print("objectiveHistory:")
for objective in objectiveHistory:
print(objective)
# Obtain the receiver-operating characteristic as a dataframe and areaUnderROC.
trainingSummary.roc.show()
print("areaUnderROC: " + str(trainingSummary.areaUnderROC))
# Set the model threshold to maximize F-Measure
fMeasure = trainingSummary.fMeasureByThreshold
maxFMeasure = fMeasure.groupBy().max('F-Measure').select('max(F-Measure)').head()
bestThreshold = fMeasure.where(fMeasure['F-Measure'] == maxFMeasure['max(F-Measure)']) \
.select('threshold').head()['threshold']
lr.setThreshold(bestThreshold)
1.2 多项逻辑回归
通过多项逻辑(softmax)回归支持多类别分类。 在多项逻辑回归中,该算法会生成K个系数集,或一个尺寸为K×J的矩阵,其中K是结果类的数量,J是特征的数量。
多项式系数可作为cofficientMatirx使用,截距可作为interceptVector使用
from pyspark.ml.classification import LogisticRegression
# Load training data
training = spark \
.read \
.format("libsvm") \
.load("data/mllib/sample_multiclass_classification_data.txt")
lr = LogisticRegression(maxIter=10, regParam=0.3, elasticNetParam=0.8)
# Fit the model
lrModel = lr.fit(training)
# Print the coefficients and intercept for multinomial logistic regression
print("Coefficients: \n" + str(lrModel.coefficientMatrix))
print("Intercept: " + str(lrModel.interceptVector))
trainingSummary = lrModel.summary
# Obtain the objective per iteration
objectiveHistory = trainingSummary.objectiveHistory
print("objectiveHistory:")
for objective in objectiveHistory:
print(objective)
# for multiclass, we can inspect metrics on a per-label basis
print("False positive rate by label:")
for i, rate in enumerate(trainingSummary.falsePositiveRateByLabel):
print("label %d: %s" % (i, rate))
print("True positive rate by label:")
for i, rate in enumerate(trainingSummary.truePositiveRateByLabel):
print("label %d: %s" % (i, rate))
print("Precision by label:")
for i, prec in enumerate(trainingSummary.precisionByLabel):
print("label %d: %s" % (i, prec))
print("Recall by label:")
for i, rec in enumerate(trainingSummary.recallByLabel):
print("label %d: %s" % (i, rec))
print("F-measure by label:")
for i, f in enumerate(trainingSummary.fMeasureByLabel()):
print("label %d: %s" % (i, f))
accuracy = trainingSummary.accuracy
falsePositiveRate = trainingSummary.weightedFalsePositiveRate
truePositiveRate = trainingSummary.weightedTruePositiveRate
fMeasure = trainingSummary.weightedFMeasure()
precision = trainingSummary.weightedPrecision
recall = trainingSummary.weightedRecall
print("Accuracy: %s\nFPR: %s\nTPR: %s\nF-measure: %s\nPrecision: %s\nRecall: %s"
% (accuracy, falsePositiveRate, truePositiveRate, fMeasure, precision, recall))
2 决策树
以下示例以LibSVM格式加载数据集,将其分为训练集和测试集,在第一个数据集上进行训练,然后对保留的测试集进行评估。 我们使用两个特征转换器准备数据。 这些帮助为标签和分类功能的索引类别,将元数据添加到决策树算法可以识别的DataFrame中。
from pyspark.ml import Pipeline
from pyspark.ml.classification import DecisionTreeClassifier
from pyspark.ml.feature import StringIndexer, VectorIndexer
from pyspark.ml.evaluation import MulticlassClassificationEvaluator
# Load the data stored in LIBSVM format as a DataFrame.
data = spark.read.format("libsvm").load("data/mllib/sample_libsvm_data.txt")
# Index labels, adding metadata to the label column.
# Fit on whole dataset to include all labels in index.
labelIndexer = StringIndexer(inputCol="label", outputCol="indexedLabel").fit(data)
# Automatically identify categorical features, and index them.
# We specify maxCategories so features with > 4 distinct values are treated as continuous.
featureIndexer =\
VectorIndexer(inputCol="features", outputCol="indexedFeatures", maxCategories=4).fit(data)
# Split the data into training and test sets (30% held out for testing)
(trainingData, testData) = data.randomSplit([0.7, 0.3])
# Train a DecisionTree model.
dt = DecisionTreeClassifier(labelCol="indexedLabel", featuresCol="indexedFeatures")
# Chain indexers and tree in a Pipeline
pipeline = Pipeline(stages=[labelIndexer, featureIndexer, dt])
# Train model. This also runs the indexers.
model = pipeline.fit(trainingData)
# Make predictions.
predictions = model.transform(testData)
# Select example rows to display.
predictions.select("prediction", "indexedLabel", "features").show(5)
# Select (prediction, true label) and compute test error
evaluator = MulticlassClassificationEvaluator(
labelCol="indexedLabel", predictionCol="prediction", metricName="accuracy")
accuracy = evaluator.evaluate(predictions)
print("Test Error = %g " % (1.0 - accuracy))
treeModel = model.stages[2]
# summary only
print(treeModel)
3.随机森林分类
以下示例以LibSVM格式加载数据集,将其分为训练集和测试集,在第一个数据集上进行训练,然后对保留的测试集进行评估。 我们使用两个特征转换器准备数据。 这些帮助为标签和分类功能的索引类别,将元数据添加到基于树的算法可以识别的DataFrame中。
from pyspark.ml import Pipeline
from pyspark.ml.classification import RandomForestClassifier
from pyspark.ml.feature import IndexToString, StringIndexer, VectorIndexer
from pyspark.ml.evaluation import MulticlassClassificationEvaluator
# Load and parse the data file, converting it to a DataFrame.
data = spark.read.format("libsvm").load("data/mllib/sample_libsvm_data.txt")
# Index labels, adding metadata to the label column.
# Fit on whole dataset to include all labels in index.
labelIndexer = StringIndexer(inputCol="label", outputCol="indexedLabel").fit(data)
# Automatically identify categorical features, and index them.
# Set maxCategories so features with > 4 distinct values are treated as continuous.
featureIndexer =\
VectorIndexer(inputCol="features", outputCol="indexedFeatures", maxCategories=4).fit(data)
# Split the data into training and test sets (30% held out for testing)
(trainingData, testData) = data.randomSplit([0.7, 0.3])
# Train a RandomForest model.
rf = RandomForestClassifier(labelCol="indexedLabel", featuresCol="indexedFeatures", numTrees=10)
# Convert indexed labels back to original labels.
labelConverter = IndexToString(inputCol="prediction", outputCol="predictedLabel",
labels=labelIndexer.labels)
# Chain indexers and forest in a Pipeline
pipeline = Pipeline(stages=[labelIndexer, featureIndexer, rf, labelConverter])
# Train model. This also runs the indexers.
model = pipeline.fit(trainingData)
# Make predictions.
predictions = model.transform(testData)
# Select example rows to display.
predictions.select("predictedLabel", "label", "features").show(5)
# Select (prediction, true label) and compute test error
evaluator = MulticlassClassificationEvaluator(
labelCol="indexedLabel", predictionCol="prediction", metricName="accuracy")
accuracy = evaluator.evaluate(predictions)
print("Test Error = %g" % (1.0 - accuracy))
rfModel = model.stages[2]
print(rfModel) # summary only
4. 梯度提升树
以下示例以LibSVM格式加载数据集,将其分为训练集和测试集,在第一个数据集上进行训练,然后对保留的测试集进行评估。 我们使用两个特征转换器准备数据。 这些帮助为标签和分类功能的索引类别,将元数据添加到基于树的算法可以识别的DataFrame中。
from pyspark.ml import Pipeline
from pyspark.ml.classification import GBTClassifier
from pyspark.ml.feature import StringIndexer, VectorIndexer
from pyspark.ml.evaluation import MulticlassClassificationEvaluator
# Load and parse the data file, converting it to a DataFrame.
data = spark.read.format("libsvm").load("data/mllib/sample_libsvm_data.txt")
# Index labels, adding metadata to the label column.
# Fit on whole dataset to include all labels in index.
labelIndexer = StringIndexer(inputCol="label", outputCol="indexedLabel").fit(data)
# Automatically identify categorical features, and index them.
# Set maxCategories so features with > 4 distinct values are treated as continuous.
featureIndexer =\
VectorIndexer(inputCol="features", outputCol="indexedFeatures", maxCategories=4).fit(data)
# Split the data into training and test sets (30% held out for testing)
(trainingData, testData) = data.randomSplit([0.7, 0.3])
# Train a GBT model.
gbt = GBTClassifier(labelCol="indexedLabel", featuresCol="indexedFeatures", maxIter=10)
# Chain indexers and GBT in a Pipeline
pipeline = Pipeline(stages=[labelIndexer, featureIndexer, gbt])
# Train model. This also runs the indexers.
model = pipeline.fit(trainingData)
# Make predictions.
predictions = model.transform(testData)
# Select example rows to display.
predictions.select("prediction", "indexedLabel", "features").show(5)
# Select (prediction, true label) and compute test error
evaluator = MulticlassClassificationEvaluator(
labelCol="indexedLabel", predictionCol="prediction", metricName="accuracy")
accuracy = evaluator.evaluate(predictions)
print("Test Error = %g" % (1.0 - accuracy))
gbtModel = model.stages[2]
print(gbtModel) # summary only
5. 多层感知器
多层感知器分类器(MLPC)是基于前馈人工神经网络的分类器。 MLPC由多层节点组成。 每一层都完全连接到网络中的下一层。 输入层中的节点代表输入数据。 所有其他节点通过输入与节点权重w和偏差b的线性组合并应用激活函数,将输入映射到输出。 对于具有K + 1层的MLPC,可以将其写成矩阵形式,如下所示:
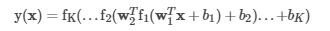
中间层中的节点使用激活函数
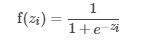
输出层中的节点使用softmax函数:
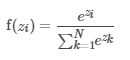
输出层中节点N的数量与类的数量相对应。 MLPC使用反向传播来学习模型。 我们使用逻辑损失函数进行优化,并使用L-BFGS作为优化例程。
from pyspark.ml.classification import MultilayerPerceptronClassifier
from pyspark.ml.evaluation import MulticlassClassificationEvaluator
# Load training data
data = spark.read.format("libsvm")\
.load("data/mllib/sample_multiclass_classification_data.txt")
# Split the data into train and test
splits = data.randomSplit([0.6, 0.4], 1234)
train = splits[0]
test = splits[1]
# specify layers for the neural network:
# input layer of size 4 (features), two intermediate of size 5 and 4
# and output of size 3 (classes)
layers = [4, 5, 4, 3]
# create the trainer and set its parameters
trainer = MultilayerPerceptronClassifier(maxIter=100, layers=layers, blockSize=128, seed=1234)
# train the model
model = trainer.fit(train)
# compute accuracy on the test set
result = model.transform(test)
predictionAndLabels = result.select("prediction", "label")
evaluator = MulticlassClassificationEvaluator(metricName="accuracy")
print("Test set accuracy = " + str(evaluator.evaluate(predictionAndLabels)))
6. 线性支持向量机
支持向量机在高维或无限维空间中构造一个超平面或一组超平面,可用于分类,回归或其他任务。 直观地,通过超平面可以实现良好的分离,该超平面与任何类别的最近训练数据点之间的距离最大(所谓的功能裕度),因为通常裕度越大,分类器的泛化误差越低。 Spark ML中的LinearSVC支持使用线性SVM进行二进制分类。 在内部,它使用OWLQN优化器优化Hinge Loss
Hinge Loss:用于训练分类器的损失函数,常被用于最大间隔分类。定义:
t为预期输出+1, -1;y为分类器的"原始输出"。当 t 和 y 符号相同(即 y 分类正确),且
时,铰接损失
。但当它们符号相反,
随 y 线性增长(分类错误)。类似的,如果
,即使 t 和 y 符号相同(正确分类,但分类间隔不足) 也会有损失。
from pyspark.ml.classification import LinearSVC
# Load training data
training = spark.read.format("libsvm").load("data/mllib/sample_libsvm_data.txt")
lsvc = LinearSVC(maxIter=10, regParam=0.1)
# Fit the model
lsvcModel = lsvc.fit(training)
# Print the coefficients and intercept for linear SVC
print("Coefficients: " + str(lsvcModel.coefficients))
print("Intercept: " + str(lsvcModel.intercept))
7. one-vs-rest / one-vs-all
OneVsRest是机器学习简化的一个示例,它提供了可以有效执行二进制分类的基本分类器,从而可以执行多分类。 它也被称为“一对多”。OneVsRest被实现为一个估计器。 对于基本分类器,它采用分类器的实例,并为k个类的每一个创建一个二进制分类问题。 训练类别i的分类器以预测标签是否为i,从而将类别i与所有其他类别区分开。 预测是通过评估每个二进制分类器来完成的,最可靠的分类器的索引将作为标签输出。
from pyspark.ml.classification import LogisticRegression, OneVsRest
from pyspark.ml.evaluation import MulticlassClassificationEvaluator
# load data file.
inputData = spark.read.format("libsvm") \
.load("data/mllib/sample_multiclass_classification_data.txt")
# generate the train/test split.
(train, test) = inputData.randomSplit([0.8, 0.2])
# instantiate the base classifier.
lr = LogisticRegression(maxIter=10, tol=1E-6, fitIntercept=True)
# instantiate the One Vs Rest Classifier.
ovr = OneVsRest(classifier=lr)
# train the multiclass model.
ovrModel = ovr.fit(train)
# score the model on test data.
predictions = ovrModel.transform(test)
# obtain evaluator.
evaluator = MulticlassClassificationEvaluator(metricName="accuracy")
# compute the classification error on test data.
accuracy = evaluator.evaluate(predictions)
print("Test Error = %g" % (1.0 - accuracy))
8.朴素贝叶斯
朴素贝叶斯分类器是一系列简单的概率分类器,基于贝叶斯定理,在每对特征之间使用强(朴素)独立性假设。朴素贝叶斯可以非常有效地训练。通过一次传递训练数据,就可以计算给定每个标签的每个特征的条件概率分布。为了进行预测,它使用贝叶斯定理来计算给定观察结果的每个标签的条件概率分布。 MLlib支持多项式朴素贝叶斯和Bernoulli朴素贝叶斯。输入数据:这些模型通常用于文档分类。在这种情况下,每个观察结果都是一个文档,每个功能都代表一个术语。特征值是术语的频率(在多项式朴素贝叶斯中),或者为零或一,表示是否在文档中找到了该术语(在Bernoulli Naive Bayes中)。特征值必须为非负数。使用可选参数“ multinomial”或“ bernoulli”(默认值为“ multinomial”)选择模型类型。对于文档分类,输入特征向量通常应为稀疏向量。由于训练数据仅使用一次,因此无需将其缓存。可以通过设置参数λ(默认值为1.0)来使用加法平滑。
from pyspark.ml.classification import NaiveBayes
from pyspark.ml.evaluation import MulticlassClassificationEvaluator
# Load training data
data = spark.read.format("libsvm") \
.load("data/mllib/sample_libsvm_data.txt")
# Split the data into train and test
splits = data.randomSplit([0.6, 0.4], 1234)
train = splits[0]
test = splits[1]
# create the trainer and set its parameters
nb = NaiveBayes(smoothing=1.0, modelType="multinomial")
# train the model
model = nb.fit(train)
# select example rows to display.
predictions = model.transform(test)
predictions.show()
# compute accuracy on the test set
evaluator = MulticlassClassificationEvaluator(labelCol="label", predictionCol="prediction",
metricName="accuracy")
accuracy = evaluator.evaluate(predictions)
print("Test set accuracy = " + str(accuracy))
二、回归
1. 线性分类
用于线性回归模型和模型摘要的接口类似于逻辑回归情况。
当通过“ l-bfgs”求解器拟合
当通过“ l-bfgs”求解器拟合具有恒定非零列的数据集而没有截距的LinearRegressionModel时,Spark MLlib为恒定非零列输出零系数。此行为与R glmnet相同,但与LIBSVM不同。
from pyspark.ml.regression import LinearRegression
# Load training data
training = spark.read.format("libsvm")\
.load("data/mllib/sample_linear_regression_data.txt")
lr = LinearRegression(maxIter=10, regParam=0.3, elasticNetParam=0.8)
# Fit the model
lrModel = lr.fit(training)
# Print the coefficients and intercept for linear regression
print("Coefficients: %s" % str(lrModel.coefficients))
print("Intercept: %s" % str(lrModel.intercept))
# Summarize the model over the training set and print out some metrics
trainingSummary = lrModel.summary
print("numIterations: %d" % trainingSummary.totalIterations)
print("objectiveHistory: %s" % str(trainingSummary.objectiveHistory))
trainingSummary.residuals.show()
print("RMSE: %f" % trainingSummary.rootMeanSquaredError)
print("r2: %f" % trainingSummary.r2)
2. 广义线性回归
与线性回归(假设输出遵循高斯分布)相反,广义线性模型(GLM)是线性模型的规范,其中响应变量Yi遵循指数分布族的某种分布。 Spark的GeneralizedLinearRegression接口可以灵活地指定GLM,可用于各种类型的预测问题,包括线性回归,泊松回归,逻辑回归等。 当前在spark.ml中,仅支持指数族分布的子集,并在下面列出。
Spark目前仅通过其GeneralizedLinearRegression接口最多支持4096个特征,如果超出此约束,将引发异常。
GLM需要指数族分布,可以以其“规范”或“自然”形式(也称为自然指数族分布)来编写。 自然指数族分布的形式为:
其中θ是parameter of interest,而τ是dispersion parameter。在GLM中,假定响应变量Yi从自然指数族分布中得出:
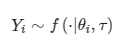
其中parameter of interestθi与期望值相关响应变量μi的值由
在这里,
由所选分布的形式定义。 GLM还允许指定链接函数,该链接函数定义了响应变量
的期望值与所谓的线性预测变量
之间的关系:
通常,选择链接函数以使
,这在感兴趣的参数θ和线性预测变量η之间产生简化的关系。在这种情况下,链接函数
被称为“规范”链接函数。
A GLM找到回归系数
最大化似然函数。
,其中 parameter of interest
与回归系数
通过
Spark的广义线性回归接口还提供了用于诊断GLM模型拟合的摘要统计信息,包括残差,p值,偏差,Akaike信息准则,和其他的。
| Family | Response Type | Supported Links |
|---|---|---|
| Gaussian | Continuous | Identity*, Log, Inverse |
| Binomial | Binary | Logit*, Probit, CLogLog |
| Poisson | Count | Log*, Identity, Sqrt |
| Gamma | Continuous | Inverse*, Idenity, Log |
| Tweedie | Zero-inflated continuous | Power link function |
from pyspark.ml.regression import GeneralizedLinearRegression
# Load training data
dataset = spark.read.format("libsvm")\
.load("data/mllib/sample_linear_regression_data.txt")
glr = GeneralizedLinearRegression(family="gaussian", link="identity", maxIter=10, regParam=0.3)
# Fit the model
model = glr.fit(dataset)
# Print the coefficients and intercept for generalized linear regression model
print("Coefficients: " + str(model.coefficients))
print("Intercept: " + str(model.intercept))
# Summarize the model over the training set and print out some metrics
summary = model.summary
print("Coefficient Standard Errors: " + str(summary.coefficientStandardErrors))
print("T Values: " + str(summary.tValues))
print("P Values: " + str(summary.pValues))
print("Dispersion: " + str(summary.dispersion))
print("Null Deviance: " + str(summary.nullDeviance))
print("Residual Degree Of Freedom Null: " + str(summary.residualDegreeOfFreedomNull))
print("Deviance: " + str(summary.deviance))
print("Residual Degree Of Freedom: " + str(summary.residualDegreeOfFreedom))
print("AIC: " + str(summary.aic))
print("Deviance Residuals: ")
summary.residuals().show()
##3 3. 决策树
from pyspark.ml import Pipeline
from pyspark.ml.regression import DecisionTreeRegressor
from pyspark.ml.feature import VectorIndexer
from pyspark.ml.evaluation import RegressionEvaluator
# Load the data stored in LIBSVM format as a DataFrame.
data = spark.read.format("libsvm").load("data/mllib/sample_libsvm_data.txt")
# Automatically identify categorical features, and index them.
# We specify maxCategories so features with > 4 distinct values are treated as continuous.
featureIndexer =\
VectorIndexer(inputCol="features", outputCol="indexedFeatures", maxCategories=4).fit(data)
# Split the data into training and test sets (30% held out for testing)
(trainingData, testData) = data.randomSplit([0.7, 0.3])
# Train a DecisionTree model.
dt = DecisionTreeRegressor(featuresCol="indexedFeatures")
# Chain indexer and tree in a Pipeline
pipeline = Pipeline(stages=[featureIndexer, dt])
# Train model. This also runs the indexer.
model = pipeline.fit(trainingData)
# Make predictions.
predictions = model.transform(testData)
# Select example rows to display.
predictions.select("prediction", "label", "features").show(5)
# Select (prediction, true label) and compute test error
evaluator = RegressionEvaluator(
labelCol="label", predictionCol="prediction", metricName="rmse")
rmse = evaluator.evaluate(predictions)
print("Root Mean Squared Error (RMSE) on test data = %g" % rmse)
treeModel = model.stages[1]
# summary only
print(treeModel)
4.随机森林回归
以下示例以LibSVM格式加载数据集,将其分为训练集和测试集,在第一个数据集上进行训练,然后对保留的测试集进行评估。 我们使用特征转换器对分类特征进行索引,将元数据添加到基于树的算法可以识别的DataFrame中。
from pyspark.ml import Pipeline
from pyspark.ml.regression import RandomForestRegressor
from pyspark.ml.feature import VectorIndexer
from pyspark.ml.evaluation import RegressionEvaluator
# Load and parse the data file, converting it to a DataFrame.
data = spark.read.format("libsvm").load("data/mllib/sample_libsvm_data.txt")
# Automatically identify categorical features, and index them.
# Set maxCategories so features with > 4 distinct values are treated as continuous.
featureIndexer =\
VectorIndexer(inputCol="features", outputCol="indexedFeatures", maxCategories=4).fit(data)
# Split the data into training and test sets (30% held out for testing)
(trainingData, testData) = data.randomSplit([0.7, 0.3])
# Train a RandomForest model.
rf = RandomForestRegressor(featuresCol="indexedFeatures")
# Chain indexer and forest in a Pipeline
pipeline = Pipeline(stages=[featureIndexer, rf])
# Train model. This also runs the indexer.
model = pipeline.fit(trainingData)
# Make predictions.
predictions = model.transform(testData)
# Select example rows to display.
predictions.select("prediction", "label", "features").show(5)
# Select (prediction, true label) and compute test error
evaluator = RegressionEvaluator(
labelCol="label", predictionCol="prediction", metricName="rmse")
rmse = evaluator.evaluate(predictions)
print("Root Mean Squared Error (RMSE) on test data = %g" % rmse)
rfModel = model.stages[1]
print(rfModel) # summary only
5. 梯度上升树回归
from pyspark.ml import Pipeline
from pyspark.ml.regression import GBTRegressor
from pyspark.ml.feature import VectorIndexer
from pyspark.ml.evaluation import RegressionEvaluator
# Load and parse the data file, converting it to a DataFrame.
data = spark.read.format("libsvm").load("data/mllib/sample_libsvm_data.txt")
# Automatically identify categorical features, and index them.
# Set maxCategories so features with > 4 distinct values are treated as continuous.
featureIndexer =\
VectorIndexer(inputCol="features", outputCol="indexedFeatures", maxCategories=4).fit(data)
# Split the data into training and test sets (30% held out for testing)
(trainingData, testData) = data.randomSplit([0.7, 0.3])
# Train a GBT model.
gbt = GBTRegressor(featuresCol="indexedFeatures", maxIter=10)
# Chain indexer and GBT in a Pipeline
pipeline = Pipeline(stages=[featureIndexer, gbt])
# Train model. This also runs the indexer.
model = pipeline.fit(trainingData)
# Make predictions.
predictions = model.transform(testData)
# Select example rows to display.
predictions.select("prediction", "label", "features").show(5)
# Select (prediction, true label) and compute test error
evaluator = RegressionEvaluator(
labelCol="label", predictionCol="prediction", metricName="rmse")
rmse = evaluator.evaluate(predictions)
print("Root Mean Squared Error (RMSE) on test data = %g" % rmse)
gbtModel = model.stages[1]
print(gbtModel) # summary only
6. 生存回归
在spark.ml中,我们实现了加速失败时间(AFT)模型,该模型是用于审查数据的参数生存回归模型。 它描述了生存时间的对数模型,因此通常称为对数线性模型,用于生存分析。 与为相同目的设计的比例风险模型不同,AFT模型更易于并行化,因为每个实例独立地对目标函数做出贡献。
给定协变量
的值,对于受试对象
= 1,…,n的随机寿命,在可能进行right-censoring的情况下,AFT模型下的似然函数为:
其中
是事件发生的指标,即是否未经审查。使用
,对数似然函数采用以下形式:
其中
是基线生存函数,而
是相应的密度函数。 最常用的AFT模型基于生存时间的Weibull分布。 寿命的Weibull分布对应于寿命对数的极值分布,并且
函数为:
方程是:
由于最小化等效于最大后验概率的负对数似然性,我们用来优化的损失函数为
。
和
的梯度函数分别为:
可以将AFT模型公式化为凸优化问题,例如,找到凸函数
的最小化器的任务,该函数取决于系数矢量
和比例参数
的对数 实现基础的优化算法是L-BFGS。 该实现与R的生存函数survreg的结果匹配。
在具有非零常量列的数据集上拟合AFTSurvivalRegressionModel且不进行截取时,Spark MLlib为非零常量列输出零系数。 此行为不同于R Survival :: survreg。
from pyspark.ml.regression import AFTSurvivalRegression
from pyspark.ml.linalg import Vectors
training = spark.createDataFrame([
(1.218, 1.0, Vectors.dense(1.560, -0.605)),
(2.949, 0.0, Vectors.dense(0.346, 2.158)),
(3.627, 0.0, Vectors.dense(1.380, 0.231)),
(0.273, 1.0, Vectors.dense(0.520, 1.151)),
(4.199, 0.0, Vectors.dense(0.795, -0.226))], ["label", "censor", "features"])
quantileProbabilities = [0.3, 0.6]
aft = AFTSurvivalRegression(quantileProbabilities=quantileProbabilities,
quantilesCol="quantiles")
model = aft.fit(training)
# Print the coefficients, intercept and scale parameter for AFT survival regression
print("Coefficients: " + str(model.coefficients))
print("Intercept: " + str(model.intercept))
print("Scale: " + str(model.scale))
model.transform(training).show(truncate=False)
7. 保序回归
等渗回归属于回归算法。 在给定有限实数集
表示观察到的响应,
要拟合的未知响应值找到最小化的函数:
对于complete order subject,x1≤x2≤…≤xnx1≤x2≤…≤xn,其中
是正权重。结果函数称为保序回归,并且是唯一的。可以将其视为在顺序限制下的最小二乘问题。从本质上讲,保序回归是最适合原始数据点的单调函数。
我们实现了一个pool adjacent violators algorithm算法,该算法使用一种使保序回归并行化的方法。训练输入是一个DataFrame,其中包含三列标签,特征和权重。此外,IsotonicRegression算法具有一个名为isotonicisotonic的可选参数,默认为true。该参数指定保序回归是保序(单调增加)还是反序(单调减少)。训练返回一个IsotonicRegressionModel,该模型可用于预测已知和未知特征的标签。等渗回归的结果被视为分段线性函数。因此,预测规则为:
- 如果预测输入与训练功能完全匹配,则返回关联的预测。 如果有多个具有相同特征的预测,则返回其中之一。 哪一个未定义(与java.util.Arrays.binarySearch相同)。
- 如果预测输入低于或高于所有训练特征,则分别返回具有最低或最高特征的预测。 如果存在多个具有相同特征的预测,则分别返回最低或最高。
- 如果预测输入介于两个训练特征之间,则将预测视为分段线性函数,并根据两个最接近特征的预测来计算内插值。 如果有多个具有相同特征的值,则使用与上一点相同的规则。
from pyspark.ml.regression import IsotonicRegression
# Loads data.
dataset = spark.read.format("libsvm")\
.load("data/mllib/sample_isotonic_regression_libsvm_data.txt")
# Trains an isotonic regression model.
model = IsotonicRegression().fit(dataset)
print("Boundaries in increasing order: %s\n" % str(model.boundaries))
print("Predictions associated with the boundaries: %s\n" % str(model.predictions))
# Makes predictions.
model.transform(dataset).show()
三、输入与输出

