序列和字母表
Bio.Alphabet.IUPAC提供Protein、DNA和RNA的基本定义
扩展:
Protein——IUPAC.protein基本类;IUPAC.extended_protein常见氨基酸类
DNA——IUPAC.unambiguous_dna基本字母;IUPAC.ambiguous_dna歧义字母;IUPAC.extended_dna修饰后的碱基
RNA——IUPAC.unambiguous_rna基本字母;IUPAC.ambiguous_rna歧义字母
定义模糊序列
Seq()可以创建一个基本的序列对象
from Bio.Alphabet import Alphabet
from Bio.Seq import Seq
myseq=Seq("AGTACACTGGT")
print(myseq)
print(myseq.alphabet)
my_seq=Seq('AGTACACTGGT', Alphabet())
print(my_seq)
print(my_seq.alphabet)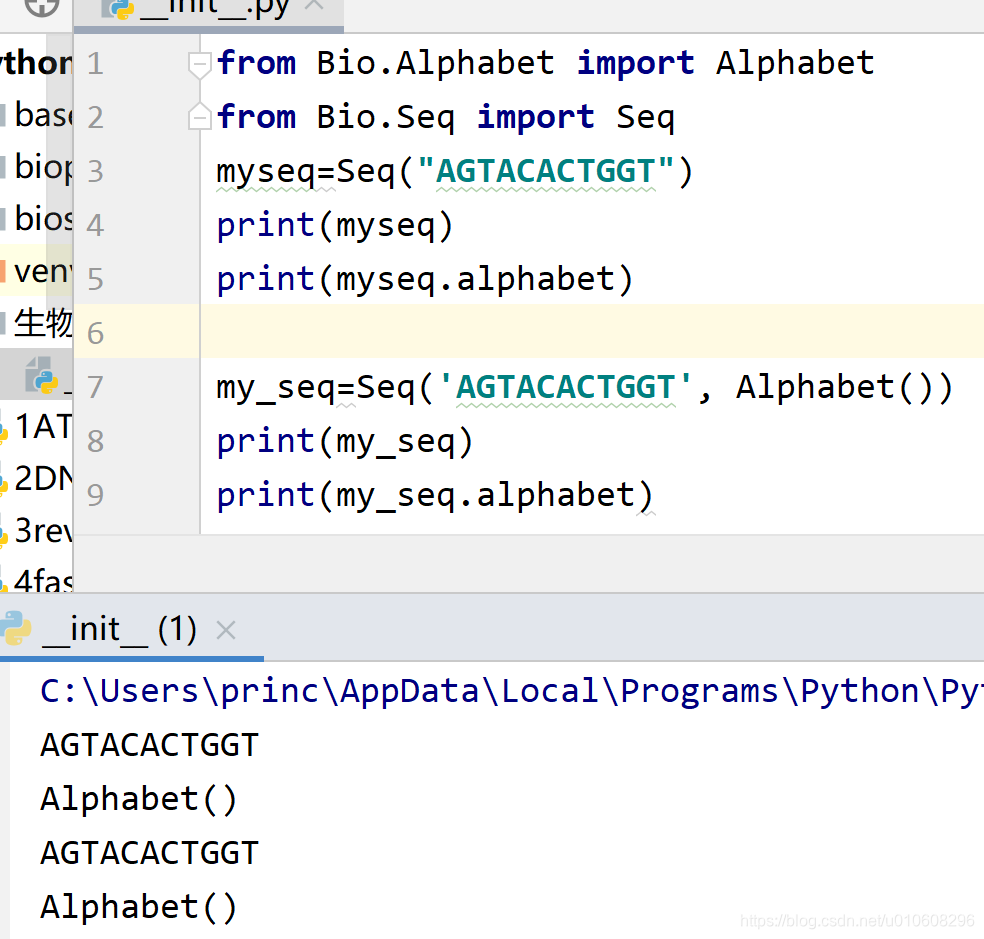
定义DNA序列
from Bio.Seq import Seq
from Bio.Alphabet import IUPAC
my_seq = Seq("AGCTGCAGCGAGCGAGC", IUPAC.unambiguous_dna)
print(my_seq)
print(my_seq.alphabet)

序列处理
迭代元素
enumerate()可以遍历序列中的元素及其下标
from Bio.Seq import Seq
from Bio.Alphabet import IUPAC
my_seq = Seq("AGTACA")
for index,letter in enumerate(my_seq): #enumerate()可以遍历序列中的元素及其下标
print(index,letter)
print(len(my_seq)) #获取长度
print(my_seq[0]) #获取序列元素
print(my_seq[4])
print(Seq("AAAAA").count("AA")) #非重叠计数统计GC含量
from Bio.Alphabet.IUPAC import IUPACUnambiguousDNA
from Bio.Seq import Seq
from Bio.SeqUtils import GC
my_seq=Seq('AGCTGA',IUPACUnambiguousDNA())
print(GC(my_seq))
切片
产生的新对象保留了原始Seq对象的字母表信息
from Bio.Alphabet.IUPAC import IUPACUnambiguousDNA
from Bio.Seq import Seq
from Bio.Alphabet import IUPAC
my_seq = Seq("AGCTGACTGACGCATGAACGATAGCA", IUPAC.unambiguous_dna)
print(my_seq[4:12])
my_seq=Seq('GACTGACG', IUPACUnambiguousDNA())
print(my_seq[4:12:3])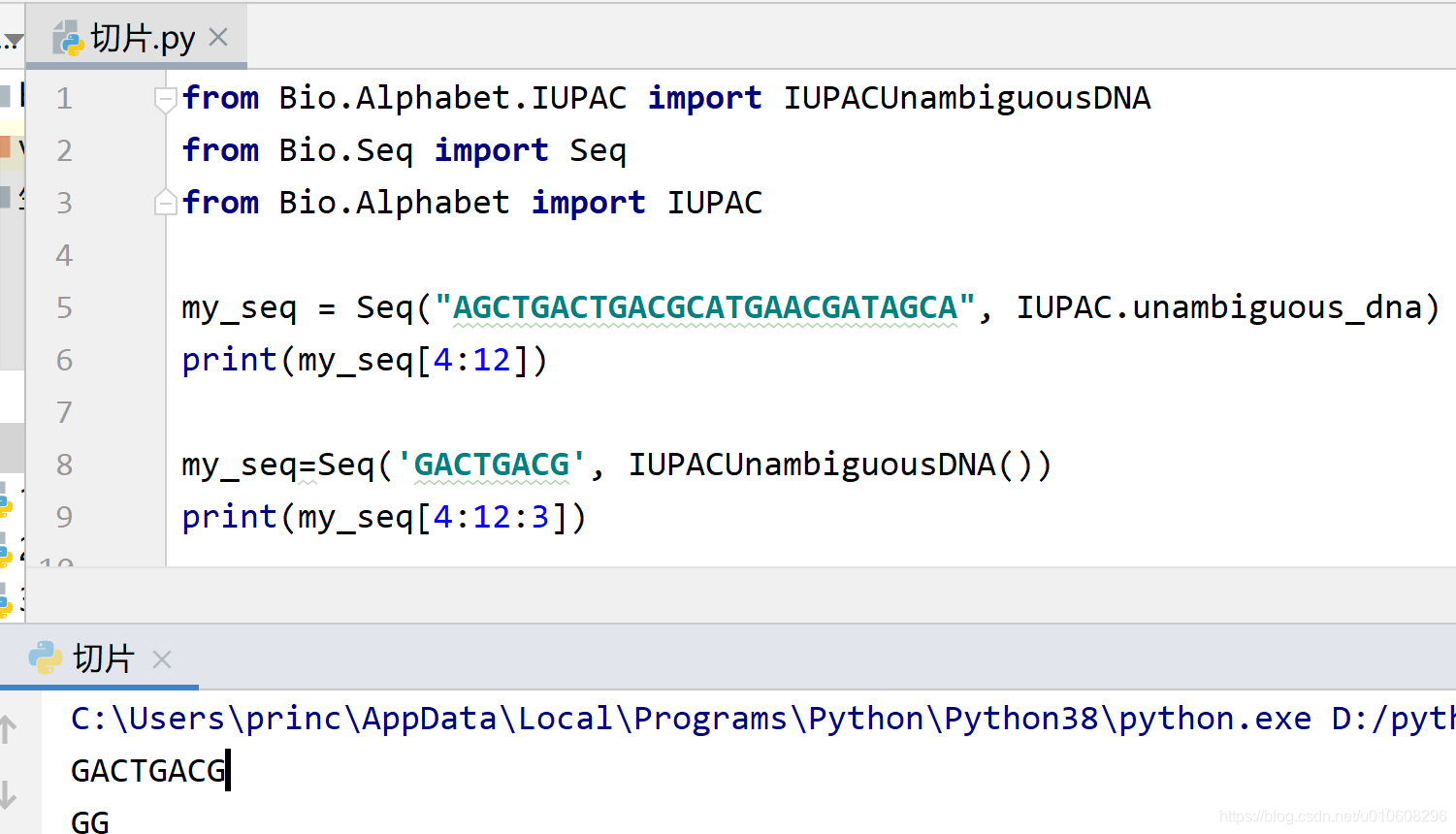
返回倒序
from Bio.Alphabet import IUPAC
from Bio.Seq import Seq
my_seq = Seq("AGCT", IUPAC.unambiguous_dna)
print(my_seq[::-1])
转换字符串
print()或%可以自动转换
from Bio.Alphabet import IUPAC
from Bio.Seq import Seq
my_seq = Seq("AGCTTAGCT", IUPAC.unambiguous_dna)
print(my_seq)
print(str(my_seq))
myseq=Seq("AGCT")
fasta=">Name\n%s\n" % myseq
print(fasta)
序列连接
相同字母表
from Bio.Alphabet import IUPAC
from Bio.Seq import Seq
dna1 = Seq("AGCT",IUPAC.unambiguous_dna)
dna2 = Seq("CGAT", IUPAC.unambiguous_dna)
dna=dna1+dna2
print(dna)
不同字母表
In[50]: from Bio.Alphabet import generic_alphabet
In[51]: protein.alphabet = generic_alphabet
In[52]: dna.alphabet = generic_alphabet
In[53]: dna + protein
Out[53]:
Seq('AGCTAGCGAAGTCCGATGEVRNAK', Alphabet())
不同字母表序列连接,必须首先将两个序列转换为通用字母表,否则会报错
ypeError: Incompatible alphabets IUPACUnambiguousDNA() and IUPACProtein()
大小写转换
from Bio.Alphabet import generic_alphabet
from Bio.Seq import Seq
my_seq = Seq("acgGATC",generic_alphabet)
print(my_seq.upper())
print(my_seq.lower())
print(generic_alphabet)
互补链和反义链
from Bio.Alphabet import IUPAC
from Bio.Seq import Seq
my_seq=Seq("GGGCCCTTT", IUPAC.unambiguous_dna)
print(my_seq) #原链
print(my_seq.complement()) #互补链
print(my_seq[::-1]) #反链
print(my_seq.reverse_complement())#反义链
生物过程模拟
转录
transcribe()将T→U转换,并调整字母表
from Bio.Seq import Seq
from Bio.Alphabet import IUPAC
coding_dna = Seq("AAAGGGTTTCCC", IUPAC.unambiguous_dna)
print(coding_dna) #原链
print(coding_dna[::-1]) #反链
template_dna = coding_dna.reverse_complement()
print(template_dna) #反义链
mRNA = coding_dna.transcribe()
print(mRNA) #转录
t_r_c=template_dna.reverse_complement().transcribe()
print(t_r_c) 
反转录
back_transcribe()从U → T的替代并伴随着字母表的变化
from Bio.Seq import Seq
from Bio.Alphabet import IUPAC
coding_dna = Seq("AAAGGGTTTCCC", IUPAC.unambiguous_dna)
mRNA = coding_dna.transcribe()
print(mRNA) #转录
print( mRNA.back_transcribe()) #反转录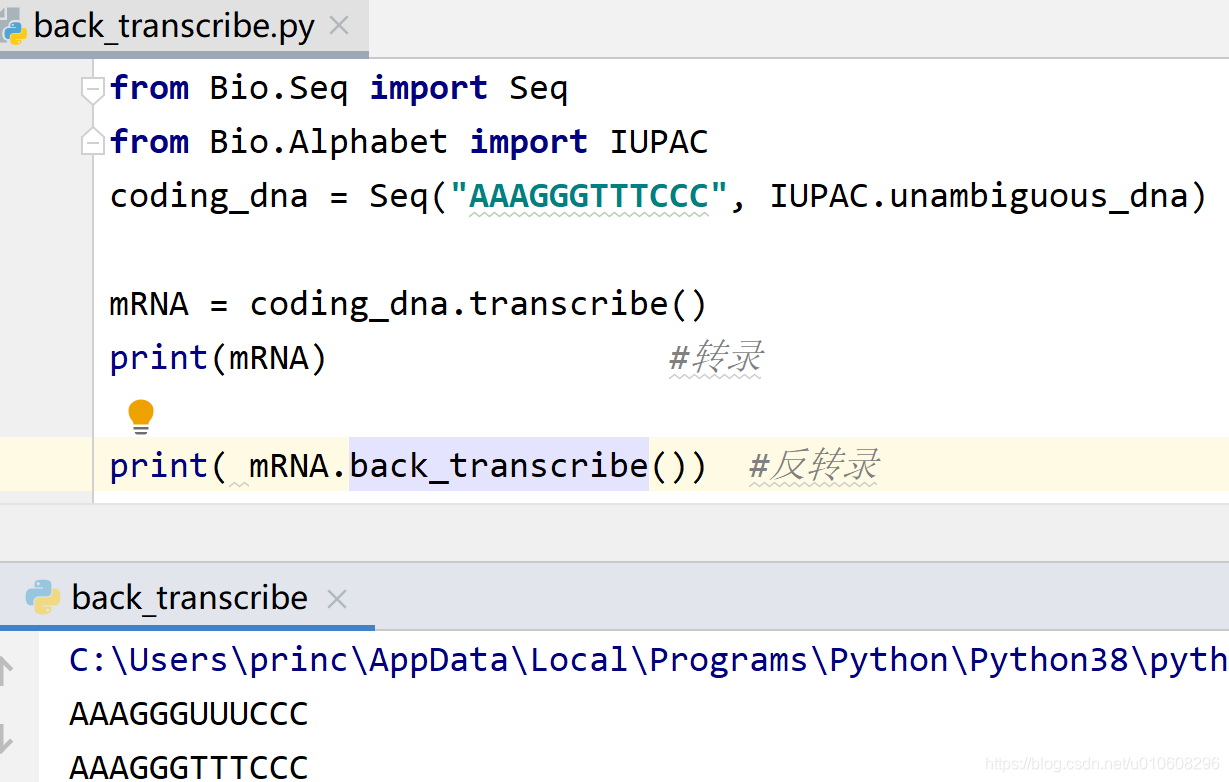
翻译
from Bio.Seq import Seq
from Bio.Alphabet import generic_dna
gene = Seq("GTGAAAAAGATGCAATCTATCGTACTCGCACTTTCCCTGGTTCTGGTCGCTCCCATGGCA" + \
"GCACAGGCTGCGGAAATTACGTTAGTCCCGTCAGTAAAATTACAGATAGGCGATCGTGAT" + \
"AATCGTGGCTATTACTGGGATGGAGGTCACTGGCGCGACCACGGCTGGTGGAAACAACAT" + \
"TATGAATGGCGAGGCAATCGCTGGCACCTACACGGACCGCCGCCACCGCCGCGCCACCAT" + \
"AAGAAAGCTCCTCATGATCATCACGGCGGTCATGGTCCAGGCAAACATCACCGCTAA",
generic_dna)
print(gene.translate(table="Bacterial"))
print(gene.translate(table="Bacterial", cds=True))
#Seq('VKKMQSIVLALSLVLVAPMAAQAAEITLVPSVKLQIGDRDNRGYYWDGGHWRDH...HR*', HasStopCodon(ExtendedIUPACProtein(), '*'))
#Seq('MKKMQSIVLALSLVLVAPMAAQAAEITLVPSVKLQIGDRDNRGYYWDGGHWRDH...HHR', ExtendedIUPACProtein())
秘密表
from Bio.Data import CodonTable
print(CodonTable.unambiguous_dna_by_name["Standard"]) #通过名字来做标识
print(CodonTable.unambiguous_dna_by_id[1]) #通过数字来做标识
Seq对象
比较
from Bio.Seq import Seq
from Bio.Alphabet import IUPAC
seq1 = Seq("AGCT", IUPAC.unambiguous_dna)
seq2 = Seq("AGCT", IUPAC.unambiguous_dna)
print(seq1 == seq2)
print(id(seq1) == id(seq2))
print(id(seq1))
print(id(seq2))
print(str(seq1) == str(seq2))

两个Seq对象,序列和字母表都时相同的,虽然seq1 == seq2 返回True,但是其实内存中这两个对象不是同一个。通过id()函数可以看到id(seq1) == id(seq2)返回False,所以在做序列比较时,可以使用str()处理后,只是以字符串比较。
可变
tomutable()
Seq对象不可变
可以使用tomutable()函数将Seq对象变为MutableSeq对象
from Bio.Seq import Seq
from Bio.Alphabet import IUPAC
my_seq = Seq("GGGTTTCCCAAA", IUPAC.unambiguous_dna)
mutable_seq = my_seq.tomutable()
print(mutable_seq)
mutable_seq[5]="G"
print(mutable_seq)
创建MutableSeq对象
可以使用toseq()将MutableSeq对象转变为Seq对象
MutableSeq对象有reverse()方法,而且各个方法直接修改MutableSeq对象本身
from Bio.Seq import Seq, MutableSeq
from Bio.Alphabet import IUPAC
mutable_seq = MutableSeq("AAACCCTTT",IUPAC.unambiguous_dna)
print(mutable_seq) # AAACCCTTT
mutable_seq[0]="T"
print(mutable_seq) # TAACCCTTT
mutable_seq.remove("T")
print(mutable_seq) # AACCCTTT 默认移除 第一个
mutable_seq.reverse()
print(mutable_seq) # TTTCCCAAA
new_seq = mutable_seq.toseq()
print(new_seq) # TTTCCCAA
print(new_seq.reverse_complement()) # TTGGGAAA

UnknownSeq对象
UnknownSeq对象可以只存储一个“N”和序列所需的长度(整数),节省内存
from Bio.Alphabet import Alphabet, IUPAC
from Bio.Seq import UnknownSeq
unk = UnknownSeq(20)
print(unk)
unk_dna = UnknownSeq(20,IUPAC.unambiguous_dna)
print(unk_dna)
print(unk)
print(unk_dna)
直接使用字符串
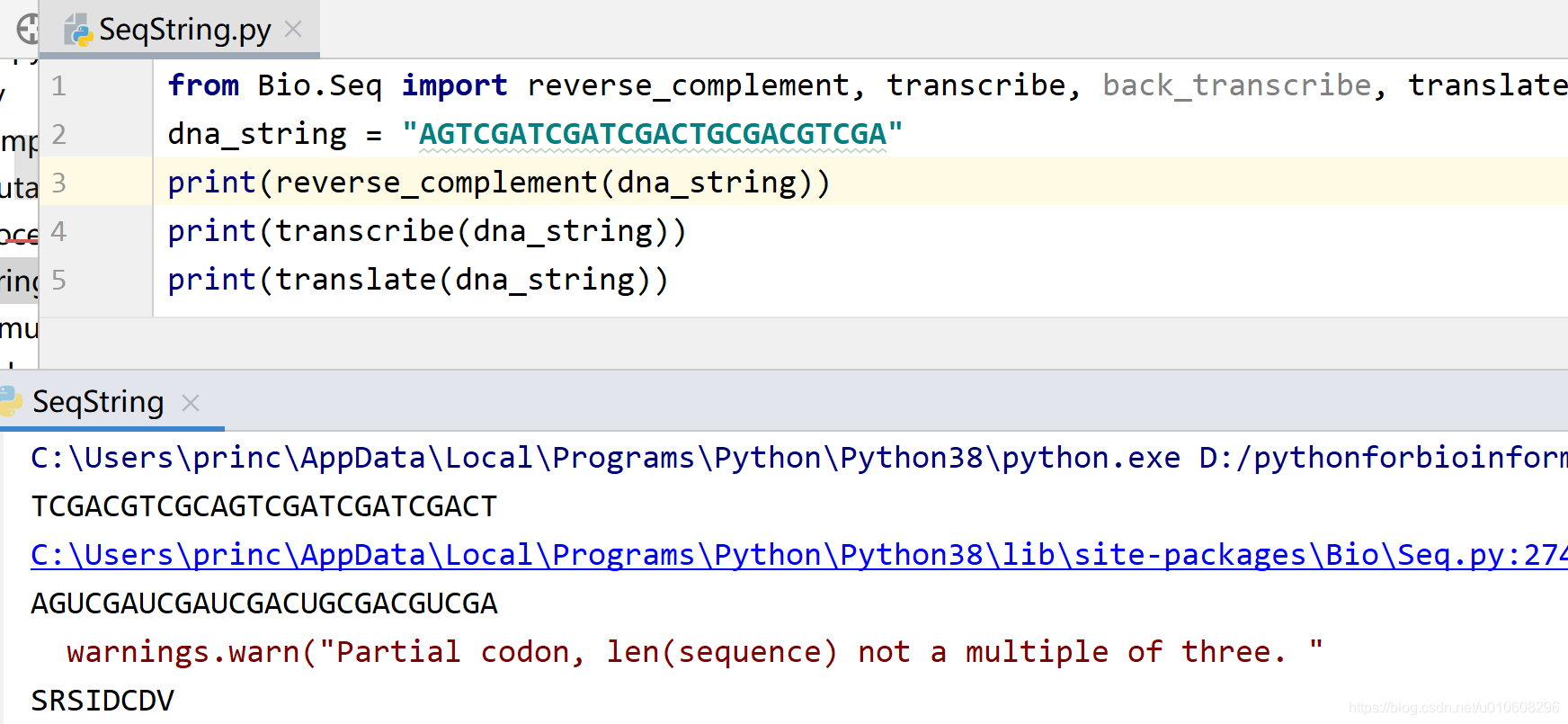
from Bio.Seq import reverse_complement, transcribe, back_transcribe, translate
dna_string = "AGTCGATCGATCGACTGCGACGTCGA"
print(reverse_complement(dna_string))
print(transcribe(dna_string))
print(translate(dna_string))