论文下载地址及翻译: 链接
网络结构:

1、Input is a four channel 3D MRI crop(4x160x192x128),the batch size to be 1 to be able to fit network into the GPU memory limits.
2、 Each green block is a ResNet-like block with the GroupNorm normalization
3、The output of the segmentation decoder has three channels (with the same spatial size as the input) followed by a sigmoid for segmentation maps of the three tumor subregions (WT、TC、ET).
4、add an additional branch(VAE) to the encoder endpoint to reconstructs the input original image into itself, and is used only during training to regularize the shared encoder,since the training dataset size is limited.
variational auto-encoder = VAE
Encoder part

1、The encoder part uses ResNet [10] blocks, where each block consists of two convolutions with normalization and ReLU, followed by additive identity skip connection
2、 For normalization, we use Group Normalization (GN) [22], which shows better than Batch Normalization(BN) performance when batch size is small (bath size of 1 in our case).
3、follow a common CNN approach to progressively downsize image dimensions by 2 and simultaneously increase feature size by 2. For downsizing we use strided convolutions. And all convolutions are 3x3x3 with initial number of filters equal to 32.
4、The encoder endpoint has size 256x20x24x16, and is 8 times spatially smaller than the input image. We decided against further downsizing to preserve more spatial content.
- 残差块,Group Normalization,在初始卷积层后面应用dropout,rate=0.2
- 下采样: strided convolution,总步长:8
- 输出:[256,16,20,32]
Decoder Part

1、每一个spatial level之间只有一个block,这和编码时候不同.
2、Each decoder level begins with upsizing
- reducing the number of features by a factor of 2 (using 1x1x1 convolutions) ,即对feature map的数量减半.
- doubling the spatial dimension (using 3D bilinear upsampling),即放大图尺寸的两倍
- followed by an addition of encoder output of the equivalent spatial level(skip connection).
3、The end of the decoder :
- has the same spatial size as the original image,
- and the number of features equal to the initial input feature size
- 然后通过1x1x1 convolution 转成 3 channels ,最后再通过 a sigmoid function来输出 three tumor subregions (WT、TC、ET)的分割图.
- 每层:1*1*1卷积是通道数减半+上采样(3D插值)+跳跃连接+一个卷积块
- 输出:[32,128,160,192]
- 最终层:1*1*1卷积输出经过sigmoid激活:[num_classes=3,128,160,192]
VAE Part

VAE分支:输入:[256,16,20,32]==>卷积==>全连接==>[256,1]
损失函数
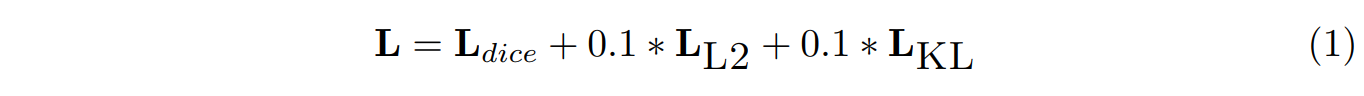

Dice loss [19]是用来描述预测和真实标签的区别

WT、TC、ET三个通道的dice loss 进行相加.
优化器
使用Adam优化器,初始化学习速率![]() ,并且会逐渐减小,公式如下:
,并且会逐渐减小,公式如下:

![]() 表示当前的epoch,
表示当前的epoch,![]() 表示总的epoch(这里使用的是300)
表示总的epoch(这里使用的是300)
正则化
1、在卷积核上使用L2正则化.
2、在初始化编码卷积后使用dropout(作者在其它地方使用并没有发现额外精度的提升)
数据的预处理和增强
1、use the largest crop size of 160x192x128 ,而原来是240x240x155(一个MR序列有155张图片,每张图片的大小为240x240)
为什么要这么裁剪?
2、将所有的输入图像进行zero mean 和 unit std
3、We apply a random (per channel) intensity shift (−0:1::0:1 ofimage std) and scale (0:9::1:1) on input image channels.
4、We also apply a random axis mirror flip (for all 3 axes) with a probability 0:5
- 在图像像素非0区域计算mean和std来normalize所有输入图像
- 在输入图像通道上应用一个随机的(每个通道)强度偏移(图像std的0:1::0:1)和放缩(0:9::1:1)
- 在3个轴上应用概率为0.5的随机的镜像翻转
总结
这么多参赛者用的方法都不完全相同,什么方法是在这个比赛中值得用的呢?根据TOP方案做分析:
- 预处理:每个图像分别做Z-Score,即对非背景(非0区域)图像减去均值,除以方差。其余的比如直方图均衡化,缩放到[0,1],用统一的均值方差Z-Score,偏移场矫正等预处理方法,TOP方案都没用。内存够用的情况下,输入的图像块尽可能大。
- 网络架构:U-Net类的编解码方式;多分支(或多head)结构;
- 后处理:Test time augmentation; model ensemble; 条件随机场(CRF)不建议用,第一名方案做了尝试,发现有的病例精度提升了,有的病例精度反而却下降了,整体没明显提升。
分类器激活函数:sigmoid,因为该网络的target不使用分类标签3个独立的类别,而使用评价的3个区域,因为这三个区域类别有交集,所以使用softmax不合理
reference
1、Brats18一参赛者对NO.1的学习总结:https://zhuanlan.zhihu.com/p/71578701
2、王国泰老师的 Test time augmentation 即测试时使用数据增强: https://arxiv.org/abs/1810.07884v1
3、一人的学习笔记: https://blog.csdn.net/gefeng1209/article/details/102755419