Article directory
"Artificial Intelligence and Machine Learning" from Prof. Manolis Kellis (Director of MIT Computational Biology)
The main content is the combination of regulatory genomics and deep convolutional networks
Since there is very little content in this part of the course I am studying, the link to the tubing is posted below (each class is one and a half hours):
Teach detailed lesson 1: Deep Learning for Regulatory Genomics - Regulator binding, Transcription Factors TFs
Taught detailed Lecture 2: Regulatory Genomics - Deep Learning in Life Sciences - Lecture 07 (Spring 2021)
Regulatory Genomics
Biological motivation of Deep CNN
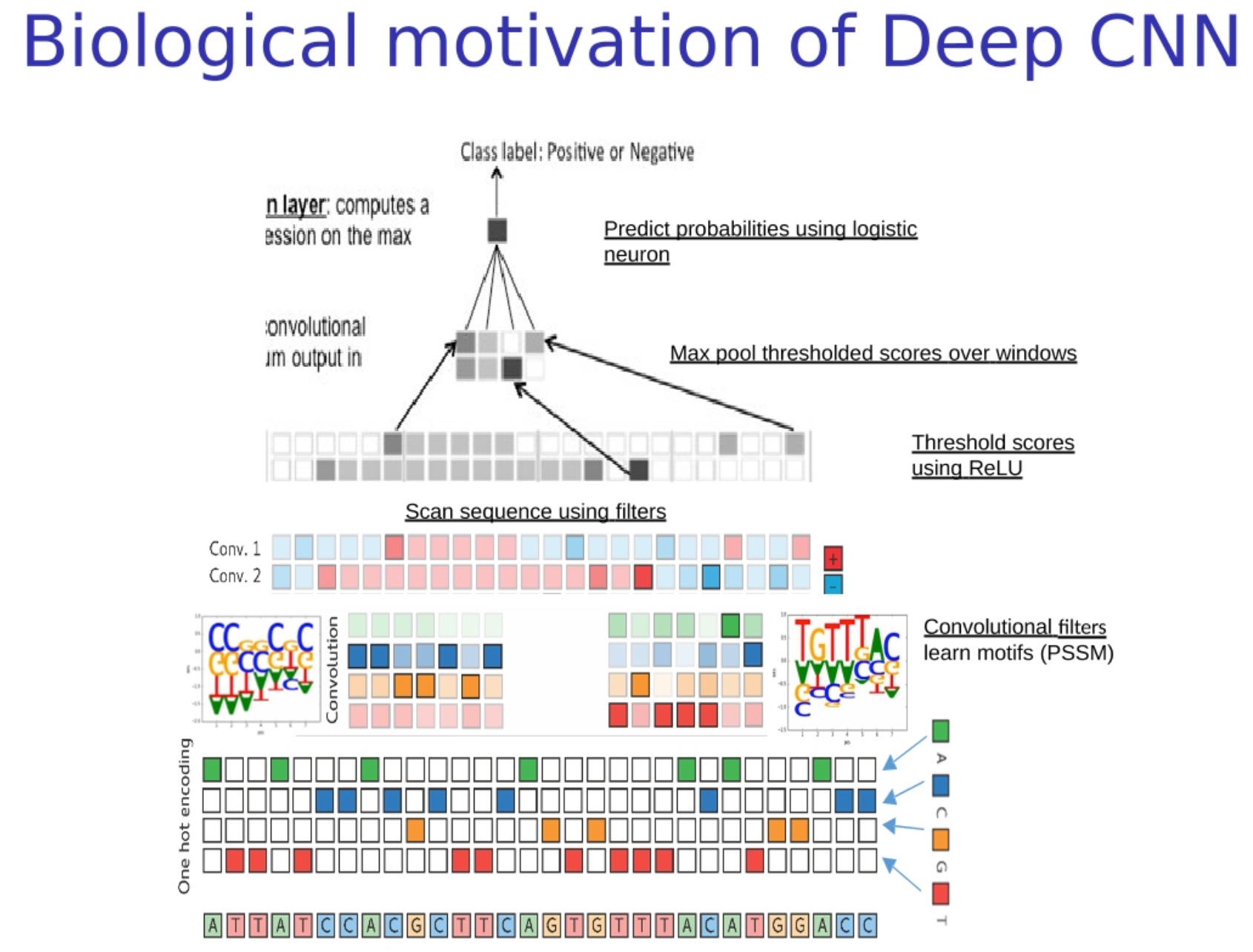
A special type of deep convolutional neural network (Deep Convolutional Neural Networks, referred to as Deep CNNs) of "deep sequence model" or "deep site-specific model", which will The filter is initialized as a Position-Specific Scoring Matrix (PSSM) or other biologically meaningful patterns (motifs). The advantage of this model is that it can use existing biological knowledge to guide the model Learning to speed up the training process and improve the accuracy of the model.
If you don't know CNN, you can see this quick primer: CNN (Convolutional Neural Network), RNN (Recurrent Neural Network), DNN (Deep Neural Network) Internals What is the difference in network structure
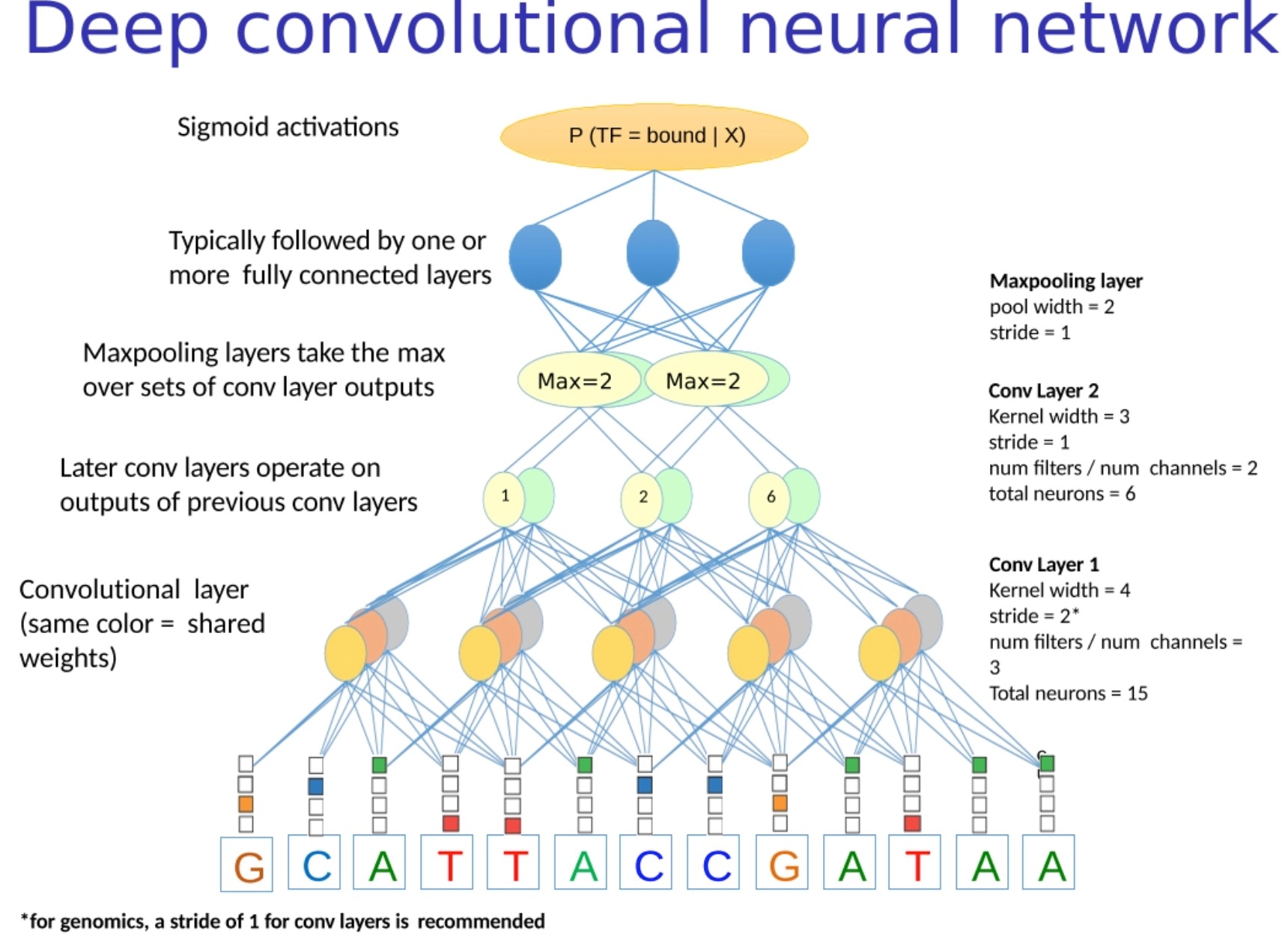
Here is the workflow for this model:
- Convert biological sequences into numerical representations : As before, we need to convert biological sequences (such as DNA) into numerical representations. The commonly used method is one-hot encoding.
- Scan sequences with biologically meaningful filters : In this step, we first initialize the filters of the convolutional layers to PSSM or other biologically meaningful patterns (motifs). These filters slide over the sequence, calculating how well the sequence matches at each position, based on the pattern that each filter corresponds to. (Unlike edge detection, the convolution kernels (filters) used here are biologically meaningful motifs)
- Thresholding by ReLU : ReLU (Rectified Linear Unit) is a common activation function that outputs 0 for negative numbers and remains unchanged for positive numbers. This operation can increase the nonlinearity of the model, allowing the model to learn more complex patterns.
- Max Pooling : This is an operation that reduces the length of a sequence while preserving key information. In convolutional neural networks, pooling layers are usually followed by convolutional layers to reduce the dimensionality of features and control overfitting.
- Predicting probabilities using logistic regression : After all the processing steps, we can use a logistic regression layer (usually a fully connected layer, plus a sigmoid activation function) to predict the class.
It is worth noting that although we initially initialize the filters to PSSM or other biologically meaningful modes, during training, the parameters of the filters are further tuned to better fit the training data. This allows us to use existing biological knowledge and learn new knowledge from data.
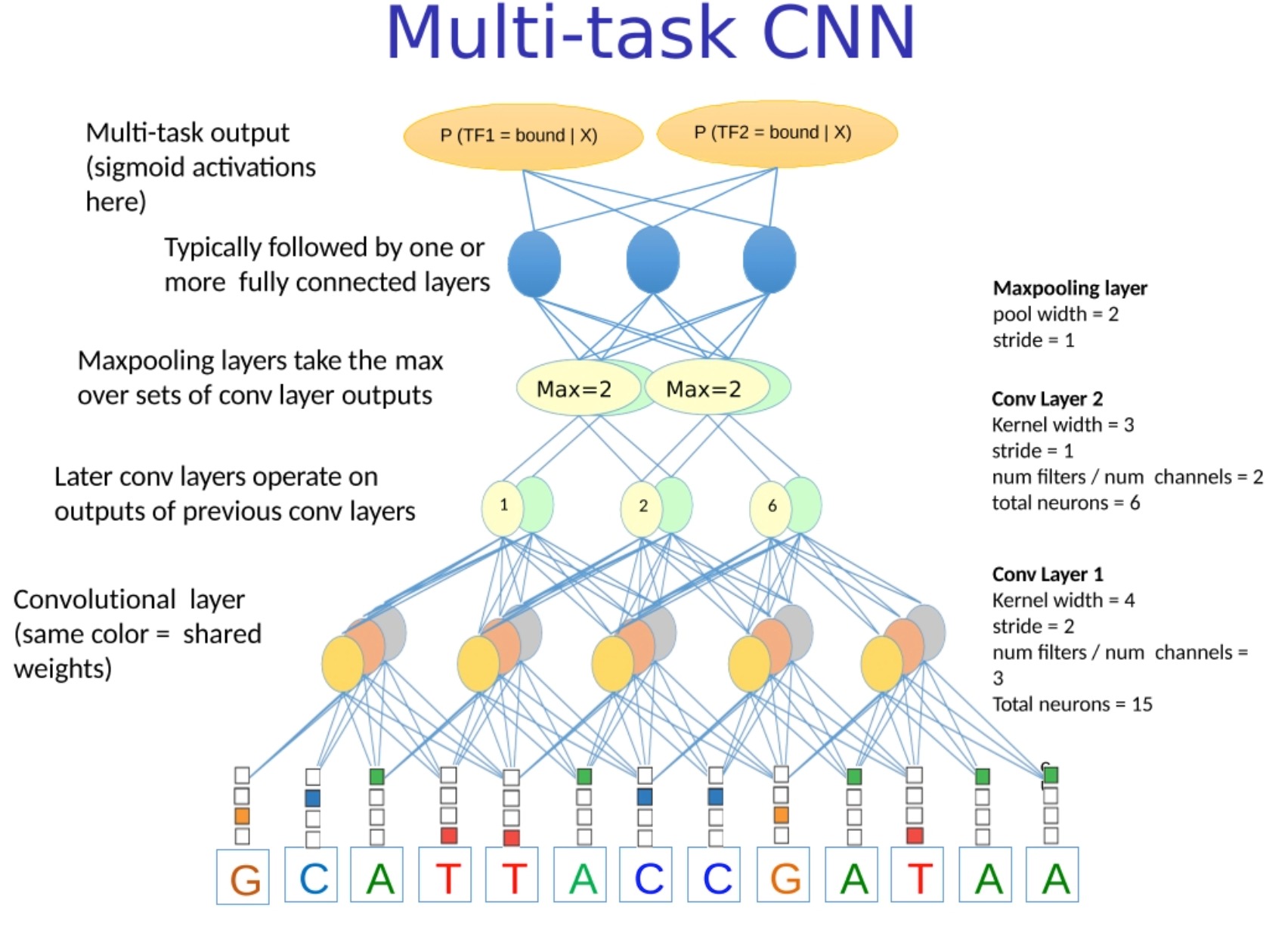
Multi-task CNN