给定一组数据:
| 样本 | 类别 | feature1 | feature2 |
|---|---|---|---|
| 0 | 1 | 1 | 5 |
| 1 | 1 | 1 | 1 |
| 2 | 1 | 2 | 4 |
| 3 | 0 | 3 | 4 |
| 4 | 0 | 4 | 5 |
第一列是类别,这里分两类,第二、三列是特征值
接下来使用Relief特征选择算法比较一下这两个特征哪个更对于分类更有作用。
第一步是计算出一个距离矩阵,比如说该矩阵的第一行是第一个样本与各个样本的距离,第二行是第二个样本与各个样本的距离,也就是说这个距离矩阵D的第i行第j列的元素
为第i个元素到第j个元素的距离。这里我们要给
赋一个较大的数,比如9999,这样在筛选猜中近邻的时候就不会选择自己了。
另外,这里的距离我们用欧几里德泛数来定义,比如样本1与样本0的距离为:
距离矩阵为:
| 9999 | 4 | 1.414 | 2.236 | 3 |
| 4 | 9999 | 3.162 | 3.606 | 5 |
| 1.414 | 3.162 | 9999 | 1 | 2.236 |
| 2.236 | 3.606 | 1 | 9999 | 1.414 |
| 3 | 5 | 2.236 | 1.414 | 9999 |
第二步是对数据表中的特征参数进行归一化处理:
利用归一化公式 对每个特征进行归一化,得
| 样本 | 类别 | feature1 | feature2 |
|---|---|---|---|
| 0 | A | 0 | 1 |
| 1 | A | 0 | 0 |
| 2 | A | ||
| 3 | B | ||
| 4 | B | 1 | 1 |
然后我们随机选择一个样本,比如选择了样本1,接下来选择样本1对于feature1的“猜中近邻”和“猜错近邻”。可以看到样本1属于类别A,而类别A中样本2与样本1的欧几里德距离最小,所以选择样本2作为样本1的关于feature1的“猜中近邻”;接下来选择“猜错近邻”,我们可以看到类别B中样本3与样本1的欧式距离最小,故选择样本3作为样本1关于feature1的“猜错近邻”,计算样本1对于feature1的相关统计量:
同理计算样本1对于feature2的相关统计量:
我们凭直觉就可以看出feature1对分类的作用大于feature2对分类的作用,然后通过上面对相关统计量的计算也可以验证这一点。上面对相关统计量的计算只是随机选择了一个样本做的计算,实际上可以随机多选择几个样本,然后将每个随机选出样本对某个特征的相关统计量求和便可以得出更令人信服的结果了。
这就是对西瓜书中式子:
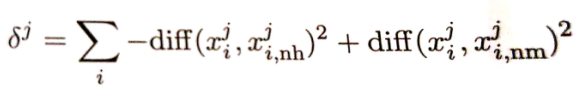 的解释。其中j代表第j个特征,i代表第i个样本,第一个diff()中第一项为被随机选中的样本i的第j个特征值,第一个diff()中第二项为样本i的“猜中近邻”的第j个特征值;第二个diff()中第一项为被随机选中的样本i的第j个特征值,第二个diff()中第二项为样本i的“猜错近邻”的第j个特征值。
的解释。其中j代表第j个特征,i代表第i个样本,第一个diff()中第一项为被随机选中的样本i的第j个特征值,第一个diff()中第二项为样本i的“猜中近邻”的第j个特征值;第二个diff()中第一项为被随机选中的样本i的第j个特征值,第二个diff()中第二项为样本i的“猜错近邻”的第j个特征值。
最后 的值越大说明第k个特征值的分类作用越好,因为最后要比较各个特征值所对应的 的大小,所以上面计算的时候要先对diff()中的特征参数进行归一化处理。
补充一下,上面所举的例子是特征值为连续的情况,当特征值为离散的情况时(也就是说特征值只用1和0表示),那么diff()中两个参数值相同则结果为1,diff()中两个参数值不同则结果为0。
python程序:
import numpy as np
from random import randrange
def relief(features, labels, times): # 传入特征矩阵,标签矩阵和随机选择的次数,因为有可能样本有很多,所以我们随机选择若干个样本来计算;这里的矩阵都是np.array
(n_samples, n_features) = np.shape(features)
delta = []
delta_features = []
delta_index = []
sample_distance = sap_distance(features) # 计算每两个样本之间的距离
new_features = normalize(features) # 对特征值归一化
"""
# 下面开始计算相关统计量,并对各个特征的相关统计量进行比较,最后返回各个特征值相关统计量从高到低的排名
# 这是将随机选取的样本代入计算出来的delta
for i in range(0, times):
randnum = randrange(0, n_samples, 1) # 生成一个随机数
one_sample = features[randnum] # 随机选择一个样本
(nearhit, nearmiss, nearhit_index, nearmiss_index) = search_near(sample_distance, labels, randnum, features) # 找出猜中近邻和猜错近邻,nearhit为猜中近邻样本的行向量
delta.append(relevant_feature(nearhit_index, nearmiss_index, new_features, randnum)) # 计算相关统计量矩阵
delta = np.asarray(delta)
for j in range(0, n_features):
delta_features.append(np.sum(delta[:, j]))
midd = list(set(delta_features))
midd.sort(reverse=True)
for p in midd:
for q in range(0, len(delta_features)):
if delta_features[q] == p:
delta_index.append(q)
return delta_index
"""
# 这是将所有样本都带入计算的delta
for i in range(0, n_samples):
(nearhit, nearmiss, nearhit_index, nearmiss_index) = search_near(sample_distance, labels, i,
features) # 找出猜中近邻和猜错近邻,nearhit为猜中近邻样本的行向量
delta.append(relevant_feature(nearhit_index, nearmiss_index, new_features, i)) # 计算相关统计量矩阵
delta = np.asarray(delta)
for j in range(0, n_features):
delta_features.append(np.sum(delta[:, j]))
midd = list(set(delta_features))
midd.sort(reverse=True)
for p in midd:
for q in range(0, len(delta_features)):
if delta_features[q] == p:
delta_index.append(q)
return delta_index
def normalize(features):
(n_samples, n_features) = np.shape(features)
print("shape=", n_samples, n_features)
fe_max = []
fe_min = []
n_deno = []
new_features = np.zeros((n_samples, n_features))
print("new_features=", new_features)
for i in range(0, n_features):
max_index = np.argmax(features[:, i])
min_index = np.argmin(features[:, i])
fe_max.append(features[max_index, i]) # 计算每一个特征的最大值
fe_min.append(features[min_index, i]) # 计算每一个特征的最小值
n_deno = np.asarray(fe_max) - np.asarray(fe_min) # 求出归一化的分母
for j in range(0, n_features):
for k in range(0, n_samples):
new_features[k, j] = (features[k, j]-fe_min[j]) / n_deno[j] # 归一化
return new_features
def sap_distance(features):
(n_samples, n_features) = np.shape(features)
distance = np.zeros((n_samples, n_samples))
for i in range(0, n_samples):
for j in range(0, n_samples):
diff_distance = features[i]-features[j]
if i == j:
distance[i, j] = 9999
else:
distance[i, j] = euclid_distance(diff_distance) # 使用欧几里德距离定义样本之间的距离
print("距离:",distance)
return distance
def euclid_distance(diff_distance):
counter = np.power(diff_distance, 2)
counter = np.sum(counter)
counter = np.sqrt(counter)
return counter
def search_near(sample_distance, labels, randnum, feartures):
(n_samples, n_features) = np.shape(feartures)
nearhit_list = []
nearmiss_list = []
hit_index = []
miss_index = []
for i in range(0, n_samples):
if labels[i] == labels[randnum]:
nearhit_list.append(sample_distance[i, randnum]) # 将距离放在一个列表里面
hit_index.append(i) # 将样本标号放在另一个列表里面
else:
nearmiss_list.append(sample_distance[i, randnum])
miss_index.append(i)
nearhit_dis_index = nearhit_list.index(min(nearhit_list)) # 算出猜中近邻
nearhit_index = hit_index[nearhit_dis_index] # 将猜中近邻的样本标号赋给nearhit_index
nearmiss_dis_index = nearmiss_list.index(min(nearmiss_list))
nearmiss_index = miss_index[nearmiss_dis_index]
nearhit = feartures[nearhit_index]
nearmiss = feartures[nearmiss_index]
return nearhit, nearmiss, nearhit_index, nearmiss_index
def relevant_feature(nearhit_index, nearmiss_index, new_features, randnum):
diff_hit = abs(new_features[nearhit_index]-new_features[randnum])
diff_miss = abs(new_features[nearmiss_index]-new_features[randnum])
delta = -np.power(diff_hit, 2)+np.power(diff_miss, 2)
return delta
out_features = np.array([[1, 5], [1, 1], [2, 4], [3, 4], [4, 5]])
labels = [1, 1, 1, 0, 0]
times = 2
features_importance = relief(out_features, labels, times)
print("排序:", features_importance)
