#逻辑斯蒂回归LogisticRegression 案例01:良/恶性乳腺癌肿瘤诊断分类(逻辑回归算法模型)
from sklearn.metrics import classification_report
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
plt.rcParams['font.sans-serif']=['SimHei']
plt.rcParams['axes.unicode_minus']=False
column_names = ['Sample code number', 'Clump Thickness', 'Uniformity of Cell Size', 'Uniformity of Cell Shape',
'Marginal Adhesion', 'Single Epithelial Cell Size', 'Bare Nuclei', 'Bland Chromatin', 'Normal Nucleoli',
'Mitoses', 'Class']
breast_cancer = pd.read_csv('breast-cancer-wisconsin.csv',names = column_names)
print(breast_cancer.head())
breast_cancer=breast_cancer.replace(to_replace='?',value=np.nan)
breast_cancer=breast_cancer.dropna(how='any')
X_train, X_test, y_train, y_test = train_test_split(breast_cancer[column_names[1:10]],
breast_cancer[column_names[10]],
test_size=0.25,
random_state=33)
print('训练样本数据集(X_train):\n{0}'.format(X_train))
print('训练样本标签集(Y_train):\n{0}'.format(y_train))
print('测试样本数据集(X_test):\n{0}'.format(X_test))
print('测试样本标签集(Y_test):\n{0}'.format(y_test))
lr=LogisticRegression()
lr.fit(X_train,y_train)
score=lr.score(X_test,y_test)
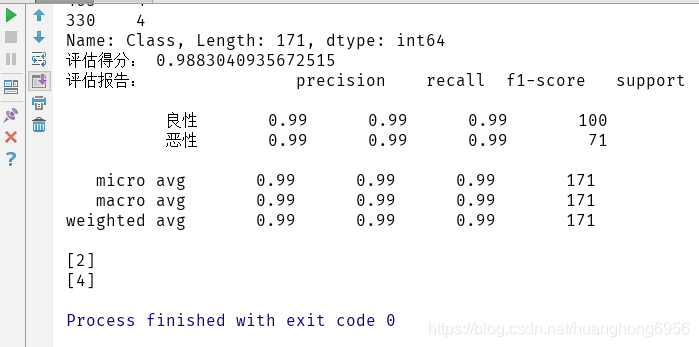
print('评估得分:',score)
result=lr.predict(X_test)
report=classification_report(y_test,result,target_names=['良性','恶性'])
print('评估报告:',report)
result=lr.predict([[2,1,1,1,2,1,3,1,1]])
print(result)
result=lr.predict([[10,7,7,3,8,5,7,4,3]])
print(result)
