由于应用需要,最近复现了一个寻优效果不过的遗传算法,算法的参考文献:
[3] Yao-Chen Chuang, Chyi-Tsong Chen, Chyi Hwang,A simple and efficient real-coded genetic algorithm for constrained optimization, Applied Soft Computing, Volume 38,2016,Pages 87-105,ISSN 1568-4946.
论文下载:https://pan.baidu.com/s/14XA1hk_tIgrfb7lgSsAziw
简单介绍一下算法操作算子和流程:由于我的问题的约束是明确的区间,而不是不等式和等式约束,所以适应度函数的计算中没有用加论文中提到的惩罚函数。
1.选择算子:
论文采取的选择算子是Ranking Selection, 其原理如下图所示:

其中0<Pr<1, 表示一个保留的比例,在选择算子的操作过程中,首先按照种群中可行解的适应度值,对可行解进行排序。排序后,用种群中前Pr*N个个体代替后Pr*N个个体。(N是种群规模),再将替换后的种群按照适应度值进行排序
2.交叉算子 (Direct Based crossover, DBX)
将排序选择后的种群分为A,B两部分


设定变异概率 ,其中
,
在集合A,B中各选取第i个个体进行交叉操作:交叉的规则为:

其中: 是两个向量的差,按照如下规则计算:

其中 是(0,1)之间的随机数。
3. 变异算子:(Dynamic Random Mutation, DRM)
按照如下的规则对种群进行变异操作:
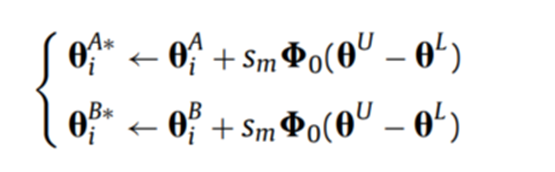
其中: 是一个与
维度相同的随机向量,向量中的每个元素介于
之间。 其中
sm的计算规则:
按照原论文中的写法,sm的计算方法如下:
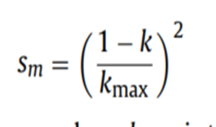
当时个人怀疑这是一种错误的写法,与论文中描述的不一致, 正确的写法应该是:,它的操作原理如下图所示:

种群的变异范围应该是随着迭代次数的增加而减小的。所以原文章中的sm计算公式是错误的。
后来证实我的怀疑是对的,在作者的另一篇文章中,将上述错误的sm的计算公式进行了改正。
算法的流程:
在传统的遗传算法的基础上,作者对算法的结构做了一些调整,无论是对进行了交叉操作还是变异操作的算子,都执行replacement operation,这样就能够对种群中的个体做到择优保留!
算法的具体流程如下图所示:

matlab代码:
clc;
clear all;
% 复现论文:[3] Yao-Chen Chuang, Chyi-Tsong Chen, Chyi Hwang,A simple and
% efficient real-coded genetic algorithm for constrained optimization,
% Applied Soft Computing, Volume 38,2016,Pages 87-105,ISSN 1568-4946.
% mode参数可设置为不同的值,选择不同的测试函数
mode = 'Schaffer';
% mode = 'Rastrigin';
% mode = 'self_define';
% mode = 'Rosenbrock';
if strcmp(mode, 'Schaffer')
figure(1)
x = -4:0.1:4;
y = -4:0.1:4;
[X,Y] = meshgrid(x,y);
% Z = 3*cos(X.*Y)+X+Y.^2;
Z = 0.5-((sin(sqrt(X.^2+Y.^2)).^2)-0.5)./(1+0.001.*(X.^2+Y.^2)).^2;
surf(X,Y,Z);
title('Schaffer Function');
xlabel('X-轴');
ylabel('Y-轴');
zlabel('Z-轴');
figure(2);
contour(X, Y, Z, 8);
title('Schaffer函数等高线');
xlabel('X-轴');
ylabel('Y-轴');
end
if strcmp(mode, 'self_define')
figure(1);
x = -4:0.1:4;
y = -4:0.1:4;
[X,Y] = meshgrid(x,y);
% Z = 100.*(Y-X.^2).^2+(1-X).^2;
Z = (cos(X.^2+Y.^2)-0.1)./(1+0.3*(X.^2+Y.^2).^2)+3;
surf(X,Y,Z);
%title('Rosen Brock valley Function');
title('Self define Function');
xlabel('X-轴');
ylabel('Y-轴');
zlabel('Z-轴');
figure(2);
contour(X, Y, Z, 8);
title('self_define函数等高线');
xlabel('X-轴');
ylabel('Y-轴');
end
if strcmp(mode, 'Rastrigin')
figure(1);
x = -4:0.1:4;
y = -4:0.1:4;
[X,Y] = meshgrid(x,y);
% Z = 100.*(Y-X.^2).^2+(1-X).^2;
%Z = (cos(X.^2+Y.^2)-0.1)./(1+0.3*(X.^2+Y.^2).^2)+3;
Z = -1.*(20+X.^2+Y.^2-10.*(cos(2.*pi.*X)+cos(2.*pi.*Y)));
surf(X,Y,Z);
%title('Rosen Brock valley Function');
title('Rastrigin Function');
xlabel('X-轴');
ylabel('Y-轴');
zlabel('Z-轴');
end
if strcmp(mode, 'Rosenbrock')
figure(1);
x_min = -2.048;
x_max = 2.048;
y_min = -2.048;
y_max = 2.048;
x = x_min:0.1:x_max;
y = y_min:0.1:y_max;
[X, Y] = meshgrid(x, y);
Z = 100.*(Y-X.^2).^2+(1-X).^2;
surf(X, Y, Z);
title('Rosenbrock Function');
xlabel('X-轴');
ylabel('Y-轴');
zlabel('Z-轴');
end
clc;
clearvars -except mode;
NP=100;
Pr = 0.10; % Ranking selection的比例
Pc=0.65; % 将Pc,Pm参数改进为自适应参数
% Pm=0.20;
G=50; % 记得改
D=2; % 变量个数
if strcmp(mode, 'Schaffer')
X_min=-4; % 变量上下界
X_max=4;
Y_min=-4;
Y_max=4;
end
if strcmp(mode, 'Rastrigin')
X_min=-4; % 变量上下界
X_max=4;
Y_min=-4;
Y_max=4;
end
if strcmp(mode, 'self_define')
X_min=-4; % 变量上下界
X_max=4;
Y_min=-4;
Y_max=4;
end
if strcmp(mode, 'Rosenbrock')
X_min=-2.048; % 变量上下界
X_max=2.048;
Y_min=-2.048;
Y_max=2.048;
end
% 产生初始种群x:
for count=1:NP % 产生初始解
temp1 = X_min+rand()*(X_max-X_min);
temp2 = Y_min+rand()*(Y_max-Y_min);
x(count,:) = [temp1,temp2];
end
save_pic_cnt = 1;
A = figure(3);
for gen=1:G
% 计算种群的适应度
pause(0.05);
if rem(gen, 2)==1
scatter(x(:,1), x(:, 2));
bias_axis = 0.5;
axis([X_min-bias_axis, X_max+bias_axis, Y_min-bias_axis, Y_max+bias_axis]);
title(['第', num2str(gen), '次迭代', ' Pr=',num2str(Pr*100), '%', ' NP=', num2str(NP)]);
xlabel('变量X');
ylabel('变量Y');
base_path = 'C:\Users\18811\Desktop\er\rosenbrock\'; % 保存图片的路径
cnt = num2str(save_pic_cnt);
tail_path = '.jpg';
frame = getframe(A);
im=frame2im(frame);
path_img = [base_path, cnt, tail_path];
% imwrite(im, path_img); % 将图片保存到设置的路径中
save_pic_cnt = save_pic_cnt + 1;
end
for count=1:NP
fitness(count)=func(x(count,:), mode);
end
fitness_max = max(fitness); % 适应度最大值
best(gen) = fitness_max; % 每一代的最优个体
fitness_min = min(fitness); % 适应度最小值
[fitness_sorted, index] = sort(fitness, 'descend'); % 对适应度降序排列
x = x(index, :); % 按照适应度值调整种群中个体的位置
proportion = NP*Pr; % Ranking selection中需要替换的个数
x(NP-proportion+1:end, :) = x(1:proportion, :); % 用好的部分解替代坏的解
% 交叉操作:directed-based crossver DBX
group_A = x(1:NP/2, :);
group_B = x(NP/2+1:end, :); % 将种群分为AB两个群体:
fitness_A = fitness_sorted(1:NP/2);
fitness_B = fitness_sorted(NP/2+1:end); % 将适应度也分为A B
for cross_cnt=1:NP/2
if rand()>Pc % 交叉概率
if all(group_A(cross_cnt, :)==group_B(cross_cnt)) || fitness_A(cross_cnt)==fitness_B(cross_cnt)
% DRM operation
group_A(cross_cnt, :) = Dynamic_random_mutation(group_A(cross_cnt, :), gen, G, X_min, X_max, Y_min, Y_max, mode);
group_B(cross_cnt, :) = Dynamic_random_mutation(group_B(cross_cnt, :), gen, G, X_min, X_max, Y_min, Y_max, mode);
else
for cnt_temp=1:D % 生成方向向量
if rand()>=0.5
D_arrow(1, cnt_temp) = group_A(cross_cnt, cnt_temp) - group_B(cross_cnt, cnt_temp);
else
D_arrow(1, cnt_temp) = 0;
end
end
S_ci = abs(fitness_A(cross_cnt)-fitness_B(cross_cnt))/(fitness_max-fitness_min); % 计算步长
group_A_new(cross_cnt, :) = group_A(cross_cnt, :) + S_ci*D_arrow;
group_B_new(cross_cnt, :) = group_B(cross_cnt, :) + S_ci*D_arrow; % 更新种群AB
% group_A_new 边界条件检查
if group_A_new(cross_cnt, 1)>X_max
group_A_new(cross_cnt, 1) = X_max;
end
if group_A_new(cross_cnt, 1)<X_min
group_A_new(cross_cnt, 1) = X_min;
end
if group_A_new(cross_cnt, 2)>Y_max
group_A_new(cross_cnt, 2) = Y_max;
end
if group_A_new(cross_cnt, 2)<Y_min
group_A_new(cross_cnt, 2) = Y_min;
end
% group_B_new 边界条件检查
if group_B_new(cross_cnt, 1)>X_max
group_B_new(cross_cnt, 1) = X_max;
end
if group_B_new(cross_cnt, 1)<X_min
group_B_new(cross_cnt, 1) = X_min;
end
if group_B_new(cross_cnt, 2)>Y_max
group_B_new(cross_cnt, 2) = Y_max;
end
if group_B_new(cross_cnt, 2)<Y_min
group_B_new(cross_cnt, 2) = Y_min;
end
% replace operation
if func(group_A_new(cross_cnt, :), mode)>fitness_A(cross_cnt)
group_A(cross_cnt, :) = group_A_new(cross_cnt, :); % replace
disp('DBX replacement');
end
if func(group_B_new(cross_cnt, :), mode)>fitness_B(cross_cnt)
group_B(cross_cnt, :) = group_B_new(cross_cnt, :); % replace
disp('DBX replacement');
end
end
else
% r_i<lambda
% DRM operation
group_A(cross_cnt, :) = Dynamic_random_mutation(group_A(cross_cnt, :), gen, G, X_min, X_max, Y_min, Y_max, mode);
group_B(cross_cnt, :) = Dynamic_random_mutation(group_B(cross_cnt, :), gen, G, X_min, X_max, Y_min, Y_max, mode);
end
end
x = [group_A; group_B]; % AB合并
end
figure(4);
plot(best);
title(['适应度进化曲线', ' Pr=',num2str(Pr*100),'%', ' NP=', num2str(NP)]);
xlabel('迭代次数');
ylabel('适应度值');
function f=func(buf, md)
if strcmp(md, 'Schaffer')
f=0.5-((sin(sqrt(buf(1).^2+buf(2).^2)).^2)-0.5)./(1+0.001.*(buf(1).^2+buf(2).^2)).^2;
end
if strcmp(md,'self_define')
% f = 100*(buf(2)-buf(1).^2).^2+(1-buf(1)).^2;
f = (cos(buf(1).^2+buf(2).^2)-0.1)./(1+0.3*(buf(1).^2+buf(2).^2).^2)+3;
end
if strcmp(md,'Rastrigin')
f = -(20+buf(1)^2+buf(2)^2-10*(cos(2*pi*buf(1))+cos(2*pi*buf(2))));
end
if strcmp(md, 'Rosenbrock')
f = 100*(buf(2)-buf(1)^2)^2+(1-buf(1))^2;
end
end
function individual_new = Dynamic_random_mutation(individual, generation_cnt, gen_max, X_min, X_max, Y_min, Y_max, mode) % 定义变异算子DRM
theta_U = [X_max, Y_max]; % 解空间的上界
theta_L = [X_min, Y_min]; % 解空间的下界
S_m = (1-generation_cnt/gen_max)^2;
temp = rand();
phi_0 = [-temp, -temp] + rand(1,2).*[2*temp, 2*temp]; % 随机扰动向量
individual_new = individual + S_m.*phi_0.*(theta_U - theta_L);
% 边界条件处理
if individual_new(1)<X_min
individual_new(1) = X_min;
end
if individual_new(1)>X_max
individual_new(1) = X_max;
end
if individual_new(2)<Y_min
individual_new(2) = Y_min;
end
if individual_new(2)>Y_max
individual_new(2) = Y_max;
end
% replacement operation
if func(individual_new, mode)<func(individual, mode)
individual_new = individual;
disp('DRM replacement');
end
end
几个运行结果:
1.Schaffer测试函数

种群分布:
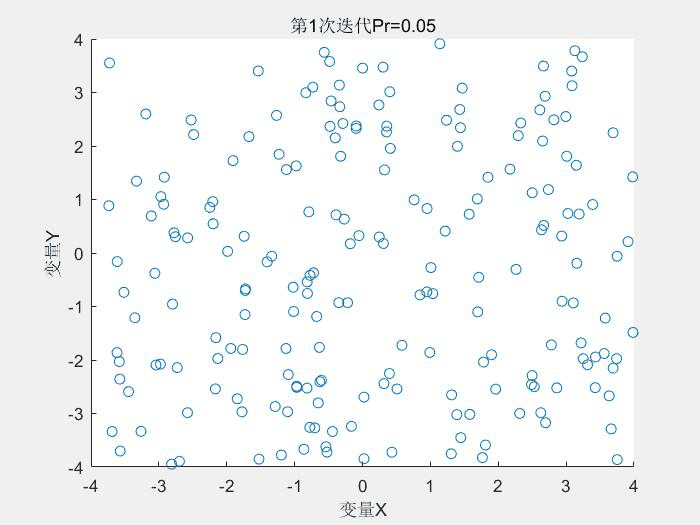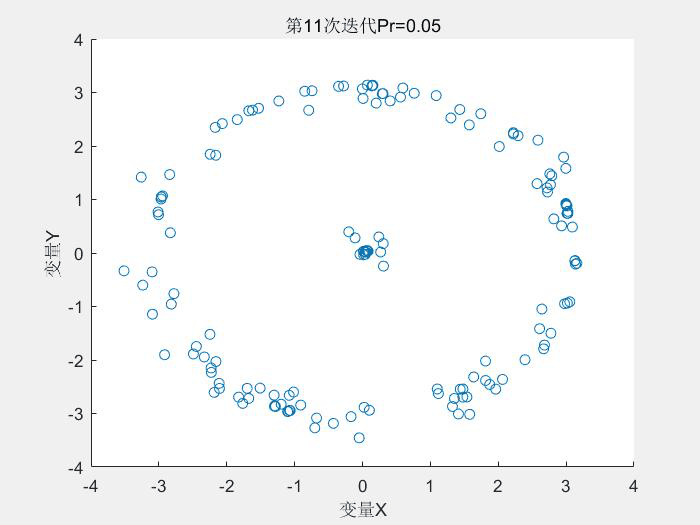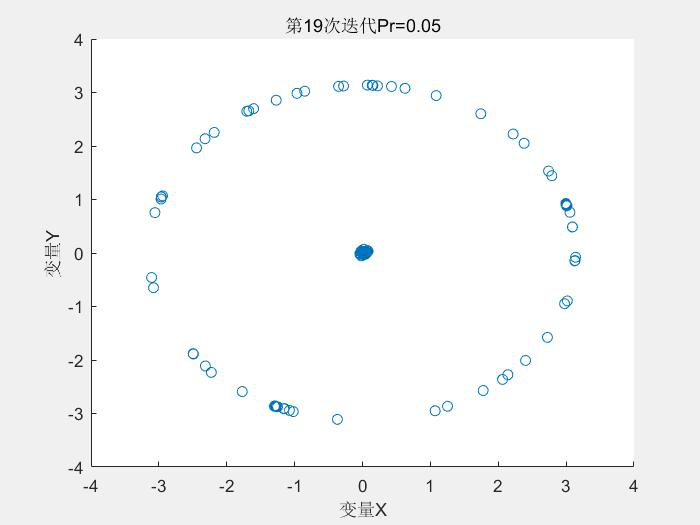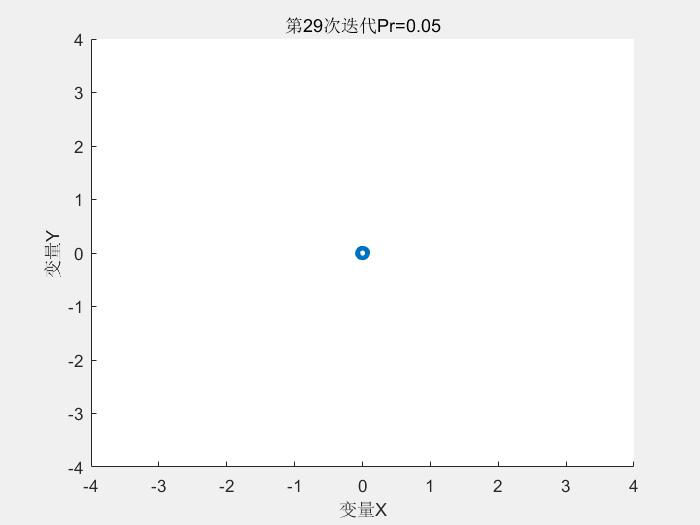

2.Rastrigin测试函数

种群分布:


可以修改代码中的参数,NP的大小,Pr的大小,观察算法的收敛情况
-----------------------------------------------分割线--------------------------------------------------------