癫痫脑电信号CNN分类实战 二
本次CNN分类实战项目没有采用Python的生物信号提取包MNE
主要内容
1数据提取
原始数据
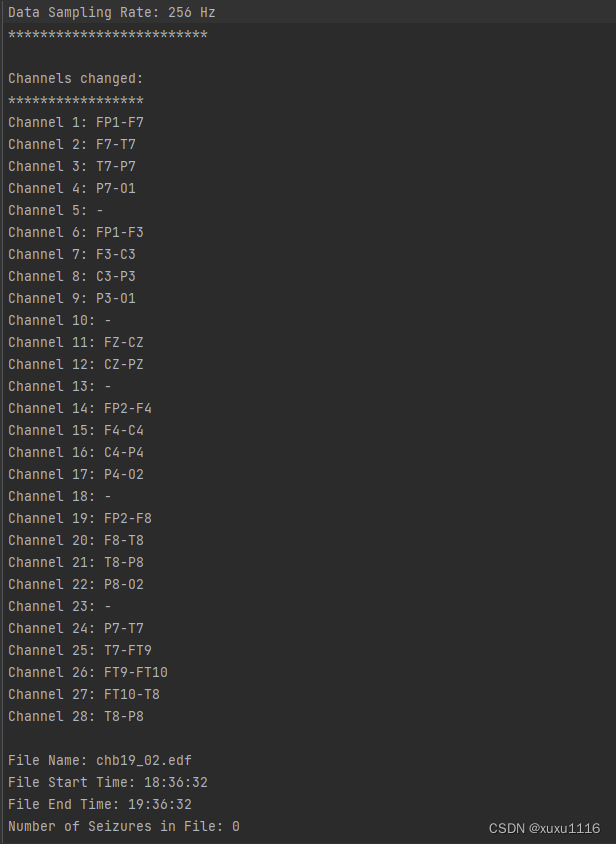
def createSpectrogram(data, S=0):
global nSpectogram
global signalsBlock
global inB
signals=np.zeros((22,59,114))
t=0
movement=int(S*256)
if(S==0):
movement=_SIZE_WINDOW_SPECTOGRAM
while data.shape[1]-(t*movement+_SIZE_WINDOW_SPECTOGRAM) > 0:
# CREAZIONE DELLO SPETROGRAMMA PER TUTTI I CANALI
for i in range(0, 22):
start = t*movement
stop = start+_SIZE_WINDOW_SPECTOGRAM
signals[i,:]=createSpec(data[i,start:stop])
if(signalsBlock is None):
signalsBlock=np.array([signals])
else:
signalsBlock=np.append(signalsBlock, [signals], axis=0)
nSpectogram=nSpectogram+1
if(signalsBlock.shape[0]==50):
saveSignalsOnDisk(signalsBlock, nSpectogram)
signalsBlock=None
# SALVATAGGIO DI SIGNALS
t = t+1
return (data.shape[1]-t*_SIZE_WINDOW_SPECTOGRAM)*-1
def createSpec(data):
fs=256
lowcut=117
highcut=123
y=butter_bandstop_filter(data, lowcut, highcut, fs, order=6)
lowcut=57
highcut=63
y=butter_bandstop_filter(y, lowcut, highcut, fs, order=6)
cutoff=1
y=butter_highpass_filter(y, cutoff, fs, order=6)
Pxx=signal.spectrogram(y, nfft=256, fs=256, return_onesided=True, noverlap=128)[2]
Pxx = np.delete(Pxx, np.s_[117:123+1], axis=0)
Pxx = np.delete(Pxx, np.s_[57:63+1], axis=0)
Pxx = np.delete(Pxx, 0, axis=0)
result=(10*np.log10(np.transpose(Pxx))-(10*np.log10(np.transpose(Pxx))).min())/(10*np.log10(np.transpose(Pxx))).ptp()
return result
def createSpectrogram(data, S=0):
global nSpectogram
global signalsBlock
global inB
signals=np.zeros((22,59,114))
t=0
movement=int(S*256)
if(S==0):
movement=_SIZE_WINDOW_SPECTOGRAM
while data.shape[1]-(t*movement+_SIZE_WINDOW_SPECTOGRAM) > 0:
# CREAZIONE DELLO SPETROGRAMMA PER TUTTI I CANALI
for i in range(0, 22):
start = t*movement
stop = start+_SIZE_WINDOW_SPECTOGRAM
signals[i,:]=createSpec(data[i,start:stop])
if(signalsBlock is None):
signalsBlock=np.array([signals])
else:
signalsBlock=np.append(signalsBlock, [signals], axis=0)
nSpectogram=nSpectogram+1
if(signalsBlock.shape[0]==50):
saveSignalsOnDisk(signalsBlock, nSpectogram)
signalsBlock=None
# SALVATAGGIO DI SIGNALS
t = t+1
return (data.shape[1]-t*_SIZE_WINDOW_SPECTOGRAM)*-1
def createArrayIntervalData(fSummary):
preictalInteval=[]
interictalInterval=[]
interictalInterval.append(PreIntData(datetime.min, datetime.max))
files=[]
firstTime=True
oldTime=datetime.min # equivalente di 0 nelle date
startTime=0
line=fSummary.readline()
endS=datetime.min
while(line):
data=line.split(':')
if(data[0]=="File Name"):
nF=data[1].strip()
s=getTime((fSummary.readline().split(": "))[1].strip())
if(firstTime):
interictalInterval[0].start=s
firstTime=False
startTime=s
while s<oldTime:#se cambia di giorno aggiungo 24 ore alla data
s=s+ timedelta(hours=24)
oldTime=s
endTimeFile=getTime((fSummary.readline().split(": "))[1].strip())
while endTimeFile<oldTime:#se cambia di giorno aggiungo 24 ore alla data
endTimeFile=endTimeFile+ timedelta(hours=24)
oldTime=endTimeFile
files.append(FileData(s, endTimeFile,nF))
for j in range(0, int((fSummary.readline()).split(':')[1])):
secSt=int(fSummary.readline().split(': ')[1].split(' ')[0])
secEn=int(fSummary.readline().split(': ')[1].split(' ')[0])
ss=s+timedelta(seconds=secSt)- timedelta(minutes=_MINUTES_OF_DATA_BETWEEN_PRE_AND_SEIZURE+_MINUTES_OF_PREICTAL)
if((len(preictalInteval)==0 or ss > endS) and ss-startTime>timedelta(minutes=20)):
ee=ss+ timedelta(minutes=_MINUTES_OF_PREICTAL)
preictalInteval.append(PreIntData(ss,ee))
endS=s+timedelta(seconds=secEn)
ss=s+timedelta(seconds=secSt)- timedelta(hours=4)
ee=s+timedelta(seconds=secEn)+ timedelta(hours=4)
if(interictalInterval[len(interictalInterval)-1].start<ss and interictalInterval[len(interictalInterval)-1].end>ee):
interictalInterval[len(interictalInterval)-1].end=ss
interictalInterval.append(PreIntData(ee, datetime.max))
else:
if(interictalInterval[len(interictalInterval)-1].start<ee):
interictalInterval[len(interictalInterval)-1].start=ee
line=fSummary.readline()
fSummary.close()
interictalInterval[len(interictalInterval)-1].end=endTimeFile
return preictalInteval, interictalInterval, files
2 滤波过滤
# Filtro taglia banda
def butter_bandstop_filter(data, lowcut, highcut, fs, order):
nyq = 0.5 * fs
low = lowcut / nyq
high = highcut / nyq
i, u = butter(order, [low, high], btype='bandstop')
y = lfilter(i, u, data)
return y
def butter_highpass_filter(data, cutoff, fs, order=5):
nyq = 0.5 * fs
normal_cutoff = cutoff / nyq
b, a = butter(order, normal_cutoff, btype='high', analog=False)
y = lfilter(b, a, data)
return y
3划分训练集和测试集
def generate_arrays_for_training(indexPat, paths, start=0, end=100):
while True:
from_=int(len(paths)/100*start)
to_=int(len(paths)/100*end)
for i in range(from_, int(to_)):
f=paths[i]
x = np.load(PathSpectogramFolder+f)
x=np.array([x])
x=x.swapaxes(0,1)
if('P' in f):
y = np.repeat([[0,1]],x.shape[0], axis=0)
else:
y =np.repeat([[1,0]],x.shape[0], axis=0)
yield(x,y)
def generate_arrays_for_predict(indexPat, paths, start=0, end=100):
while True:
from_=int(len(paths)/100*start)
to_=int(len(paths)/100*end)
for i in range(from_, int(to_)):
f=paths[i]
x = np.load(PathSpectogramFolder+f)
x=np.array([x])
x=x.swapaxes(0,1)
yield(x)
2 CNN建模
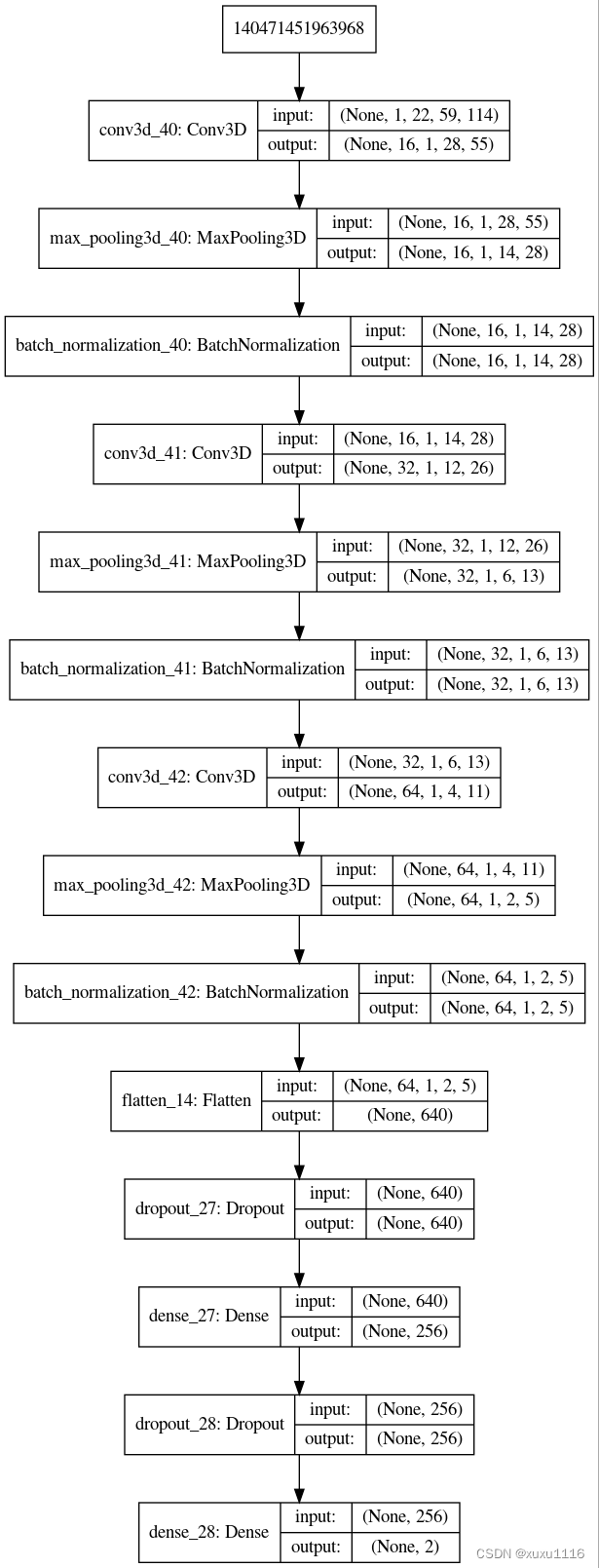
def createModel():
input_shape=(1, 22, 59, 114)
model = Sequential()
#C1
model.add(Conv3D(16, (22, 5, 5), strides=(1, 2, 2), padding='valid',activation='relu',data_format= "channels_first", input_shape=input_shape))
model.add(keras.layers.MaxPooling3D(pool_size=(1, 2, 2),data_format= "channels_first", padding='same'))
model.add(BatchNormalization())
#C2
model.add(Conv3D(32, (1, 3, 3), strides=(1, 1,1), padding='valid',data_format= "channels_first", activation='relu'))#incertezza se togliere padding
model.add(keras.layers.MaxPooling3D(pool_size=(1,2, 2),data_format= "channels_first", ))
model.add(BatchNormalization())
#C3
model.add(Conv3D(64, (1,3, 3), strides=(1, 1,1), padding='valid',data_format= "channels_first", activation='relu'))#incertezza se togliere padding
model.add(keras.layers.MaxPooling3D(pool_size=(1,2, 2),data_format= "channels_first", ))
model.add(BatchNormalization())
model.add(Flatten())
model.add(Dropout(0.5))
model.add(Dense(256, activation='sigmoid'))
model.add(Dropout(0.5))
model.add(Dense(2, activation='softmax'))
opt_adam = keras.optimizers.Adam(lr=0.00001, beta_1=0.9, beta_2=0.999, epsilon=1e-08, decay=0.0)
model.compile(loss='categorical_crossentropy', optimizer=opt_adam, metrics=['accuracy'])
return model
def main():
print("START")
if not os.path.exists(OutputPathModels):
os.makedirs(OutputPathModels)
loadParametersFromFile("PARAMETERS_CNN.txt")
#callback=EarlyStopping(monitor='val_acc', min_delta=0, patience=0, verbose=0, mode='auto', baseline=None)
callback=EarlyStoppingByLossVal(monitor='val_acc', value=0.975, verbose=1, lower=False)
print("Parameters loaded")
for indexPat in range(0, len(patients)):
print('Patient '+patients[indexPat])
if not os.path.exists(OutputPathModels+"ModelPat"+patients[indexPat]+"/"):
os.makedirs(OutputPathModels+"ModelPat"+patients[indexPat]+"/")
loadSpectogramData(indexPat)
print('Spectograms data loaded')
result='Patient '+patients[indexPat]+'\n'
result='Out Seizure, True Positive, False Positive, False negative, Second of Inter in Test, Sensitivity, FPR \n'
for i in range(0, nSeizure):
print('SEIZURE OUT: '+str(i+1))
print('Training start')
model = createModel()
filesPath=getFilesPathWithoutSeizure(i, indexPat)
model.fit_generator(generate_arrays_for_training(indexPat, filesPath, end=75), #end=75),#It take the first 75%
validation_data=generate_arrays_for_training(indexPat, filesPath, start=75),#start=75), #It take the last 25%
#steps_per_epoch=10000, epochs=10)
steps_per_epoch=int((len(filesPath)-int(len(filesPath)/100*25))),#*25),
validation_steps=int((len(filesPath)-int(len(filesPath)/100*75))),#*75),
verbose=2,
epochs=300, max_queue_size=2, shuffle=True, callbacks=[callback])# 100 epochs è meglio #aggiungere criterio di stop in base accuratezza
print('Training end')
print('Testing start')
filesPath=interictalSpectograms[i]
interPrediction=model.predict_generator(generate_arrays_for_predict(indexPat, filesPath), max_queue_size=4, steps=len(filesPath))
filesPath=preictalRealSpectograms[i]
preictPrediction=model.predict_generator(generate_arrays_for_predict(indexPat, filesPath), max_queue_size=4, steps=len(filesPath))
print('Testing end')
# Creates a HDF5 file
model.save(OutputPathModels+"ModelPat"+patients[indexPat]+"/"+'ModelOutSeizure'+str(i+1)+'.h5')
print("Model saved")
#to plot the model
#plot_model(model, to_file="CNNModel", show_shapes=True, show_layer_names=True)
if not os.path.exists(OutputPathModels+"OutputTest"+"/"):
os.makedirs(OutputPathModels+"OutputTest"+"/")
np.savetxt(OutputPathModels+"OutputTest"+"/"+"Int_"+patients[indexPat]+"_"+str(i+1)+".csv", interPrediction, delimiter=",")
np.savetxt(OutputPathModels+"OutputTest"+"/"+"Pre_"+patients[indexPat]+"_"+str(i+1)+".csv", preictPrediction, delimiter=",")
secondsInterictalInTest=len(interictalSpectograms[i])*50*30#50 spectograms for file, 30 seconds for each spectogram
acc=0#accumulator
fp=0
tp=0
fn=0
lastTenResult=list()
for el in interPrediction:
if(el[1]>0.5):
acc=acc+1
lastTenResult.append(1)
else:
lastTenResult.append(0)
if(len(lastTenResult)>10):
acc=acc-lastTenResult.pop(0)
if(acc>=8):
fp=fp+1
lastTenResult=list()
acc=0
lastTenResult=list()
for el in preictPrediction:
if(el[1]>0.5):
acc=acc+1
lastTenResult.append(1)
else:
lastTenResult.append(0)
if(len(lastTenResult)>10):
acc=acc-lastTenResult.pop(0)
if(acc>=8):
tp=tp+1
else:
if(len(lastTenResult)==10):
fn=fn+1
sensitivity=tp/(tp+fn)
FPR=fp/(secondsInterictalInTest/(60*60))
result=result+str(i+1)+','+str(tp)+','+str(fp)+','+str(fn)+','+str(secondsInterictalInTest)+','
result=result+str(sensitivity)+','+str(FPR)+'\n'
print('True Positive, False Positive, False negative, Second of Inter in Test, Sensitivity, FPR')
print(str(tp)+','+str(fp)+','+str(fn)+','+str(secondsInterictalInTest)+','+str(sensitivity)+','+str(FPR))
with open(OutputPath, "a+") as myfile:
myfile.write(result)

项目训练所需的环境
python3.6.5
tensorflow1.10
cuda10.0.130
cudnn7.6.5
显卡2080TI
需要代码直接私信我