文章主要参考了《matlab智能算法30例》,仅供学习交流使用。
%% 免疫优化算法在物流配送中心选址中的应用
%% 清空环境
clc
clear
%% 算法基本参数
sizepop=50; % 种群规模
overbest=10; % 记忆库容量
MAXGEN=100; % 迭代次数
pcross=0.5; % 交叉概率
pmutation=0.4; % 变异概率
ps=0.95; % 多样性评价参数
length=6; % 配送中心数
M=sizepop+overbest;
%% step1 识别抗原,将种群信息定义为一个结构体
individuals = struct('fitness',zeros(1,M), 'concentration',zeros(1,M),'excellence',zeros(1,M),'chrom',[]);
%% step2 产生初始抗体群
individuals.chrom = popinit(M,length);
trace=[]; %记录每代最个体优适应度和平均适应度
%% 迭代寻优
for iii=1:MAXGEN
%% step3 抗体群多样性评价
for i=1:M
individuals.fitness(i) = fitness(individuals.chrom(i,:)); % 抗体与抗原亲和度(适应度值)计算
individuals.concentration(i) = concentration(i,M,individuals); % 抗体浓度计算
end
% 综合亲和度和浓度评价抗体优秀程度,得出繁殖概率
individuals.excellence = excellence(individuals,M,ps);
% 记录当代最佳个体和种群平均适应度
[best,index] = min(individuals.fitness); % 找出最优适应度
bestchrom = individuals.chrom(index,:); % 找出最优个体
average = mean(individuals.fitness); % 计算平均适应度
trace = [trace;best,average]; % 记录
%% step4 根据excellence,形成父代群,更新记忆库(加入精英保留策略,可由s控制)
bestindividuals = bestselect(individuals,M,overbest); % 更新记忆库
individuals = bestselect(individuals,M,sizepop); % 形成父代群
%% step5 选择,交叉,变异操作,再加入记忆库中抗体,产生新种群
individuals = select(individuals,sizepop); % 选择
individuals.chrom = Cross(pcross,individuals.chrom,sizepop,length); % 交叉
individuals.chrom = mutation(pmutation,individuals.chrom,sizepop,length); % 变异
individuals = incorporate(individuals,sizepop,bestindividuals,overbest); % 加入记忆库中抗体
end
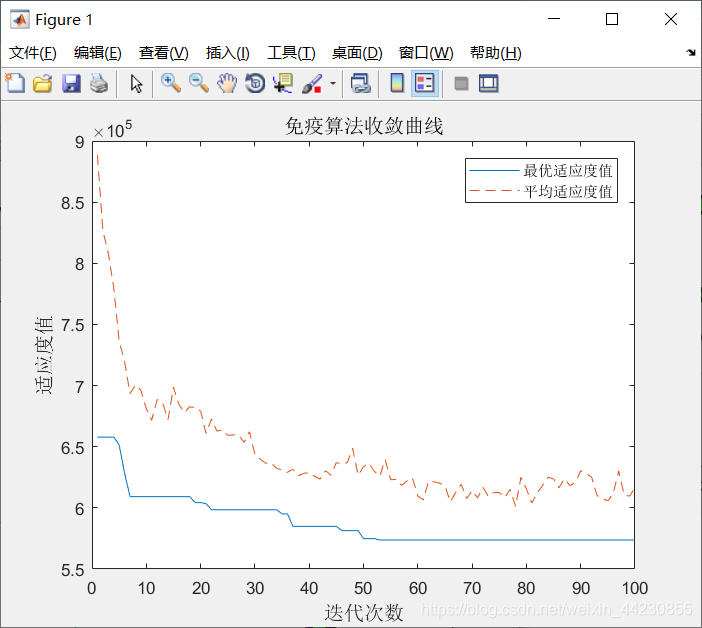
%% 画出免疫算法收敛曲线
figure(1)
plot(trace(:,1));
hold on
plot(trace(:,2),'--');
legend('最优适应度值','平均适应度值')
title('免疫算法收敛曲线','fontsize',12)
xlabel('迭代次数','fontsize',12)
ylabel('适应度值','fontsize',12)
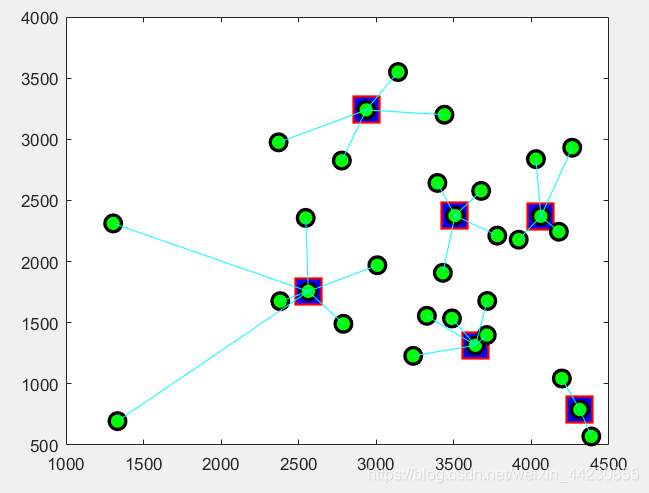
%% 画出配送中心选址图
%城市坐标
city_coordinate=[1304,2312;3639,1315;4177,2244;3712,1399;3488,1535;3326,1556;3238,1229;4196,1044;4312,790;4386,570;
3007,1970;2562,1756;2788,1491;2381,1676;1332,695;3715,1678;3918,2179;4061,2370;3780,2212;3676,2578;
4029,2838;4263,2931;3429,1908;3507,2376;3394,2643;3439,3201;2935,3240;3140,3550;2545,2357;2778,2826;2370,2975];
carge=[20,90,90,60,70,70,40,90,90,70,60,40,40,40,20,80,90,70,100,50,50,50,80,70,80,40,40,60,70,50,30];
%找出最近配送点
for i=1:31
distance(i,:)=dist(city_coordinate(i,:),city_coordinate(bestchrom,:)');
end
[a,b]=min(distance');
index=cell(1,length);
for i=1:length
%计算各个派送点的地址
index{i}=find(b==i);
end
figure(2)
title('最优规划派送路线')
cargox=city_coordinate(bestchrom,1);
cargoy=city_coordinate(bestchrom,2);
plot(cargox,cargoy,'rs','LineWidth',2,...
'MarkerEdgeColor','r',...
'MarkerFaceColor','b',...
'MarkerSize',20)
hold on
plot(city_coordinate(:,1),city_coordinate(:,2),'o','LineWidth',2,...
'MarkerEdgeColor','k',...
'MarkerFaceColor','g',...
'MarkerSize',10)
for i=1:31
x=[city_coordinate(i,1),city_coordinate(bestchrom(b(i)),1)];
y=[city_coordinate(i,2),city_coordinate(bestchrom(b(i)),2)];
plot(x,y,'c');hold on
end
function fit=fitness(individual)
%计算个体适应度
%individual input 个体
%fit output 适应度值
%城市坐标
city_coordinate=[1304,2312;3639,1315;4177,2244;3712,1399;3488,1535;3326,1556;
3238,1229;4196,1044;4312,790;4386,570;3007,1970;2562,1756;
2788,1491;2381,1676;1332,695;3715,1678;3918,2179;4061,2370;
3780,2212;3676,2578;4029,2838;4263,2931;3429,1908;3507,2376;
3394,2643;3439,3201;2935,3240;3140,3550;2545,2357;2778,2826;
2370,2975];
%货物量
carge=[20,90,90,60,70,70,40,90,90,70,60,40,40,40,20,80,90,70,100,50,50,50,80,70,80,40,40,60,70,50,30];
%找出最近配送点
for i=1:31
%dist函数就是欧式距离加权函数
distance(i,:)=dist(city_coordinate(i,:),city_coordinate(individual,:)');
end
[a,b]=min(distance');
%计算费用
for i=1:31
expense(i)=carge(i)*a(i);
end
fit=sum(expense)+4.0e+4*length(find(a>3000));
end
function resemble=similar(individual1,individual2)
%计算个体individual1与individual2的相似度
%individual1,individual2 input 两个个体
%resemble output 相似度
k=zeros(1,length(individual1));
for i=1:length(individual1)
if find(individual1(i)==individual2)
k(i)=1;
end
end
resemble=sum(k)/length(individual1);
end
function concentration = concentration(i,M,individuals)
% 计算个体浓度值
% i input 第i个抗体
% M input 种群规模
% individuals input 个体
% concentration output 浓度值
concentration=0;
for j=1:M
xsd=similar(individuals.chrom(i,:),individuals.chrom(j,:)); % 第i个体与种群个体间的相似度
% 相似度大于阀值
if xsd>0.7
concentration=concentration+1;
end
end
concentration=concentration/M;
end
function exc = excellence(individuals,M,ps)
%计算个体繁殖概率
%individuals input 种群
%M input 种群规模
%ps input 多样性评价参数
%exc output 繁殖概率
fit = 1./individuals.fitness;
sumfit = sum(fit);
con = individuals.concentration;
sumcon = sum(con);
for i=1:M
exc(i)=fit(i)/sumfit*ps+con(i)/sumcon*(1-ps);
end
end
function ret=Select(individuals,sizepop)
% 轮盘赌选择
% individuals input : 种群信息
% sizepop input : 种群规模
% ret output : 选择后得到的种群
excellence=individuals.excellence;
pselect=excellence./sum(excellence);
% 事实上 pselect = excellence;
index=[];
for i=1:sizepop % 转sizepop次轮盘
pick=rand;
while pick==0
pick=rand;
end
for j=1:sizepop
pick=pick-pselect(j);
if pick<0
index=[index j];
break; % 寻找落入的区间,此次转轮盘选中了染色体j
end
end
end
% 注意:在转sizepop次轮盘的过程中,有可能会重复选择某些染色体
individuals.chrom=individuals.chrom(index,:);
individuals.fitness=individuals.fitness(index);
individuals.concentration=individuals.concentration(index);
individuals.excellence=individuals.excellence(index);
ret=individuals;
end
function ret=Mutation(pmutation,chrom,sizepop,length1)
% 变异操作
% pmutation input : 变异概率
% chrom input : 抗体群
% sizepop input : 种群规模
% iii input : 进化代数
% MAXGEN input : 最大进化代数
% length1 input : 抗体长度
% ret output : 变异得到的抗体群
% 每一轮for循环中,可能会进行一次变异操作,染色体是随机选择的,变异位置也是随机选择的
for i=1:sizepop
% 变异概率
pick=rand;
while pick==0
pick=rand;
end
index=unidrnd(sizepop);
% 判断是否变异
if pick>pmutation
continue;
end
pos=unidrnd(length1);
while pos==1
pos=unidrnd(length1);
end
nchrom=chrom(index,:);
nchrom(pos)=unidrnd(31);
while length(unique(nchrom))==(length1-1)
nchrom(pos)=unidrnd(31);
end
flag=test(nchrom);
if flag==1
chrom(index,:)=nchrom;
end
end
ret=chrom;
end
function newindividuals = incorporate(individuals,sizepop,bestindividuals,overbest)
% 将记忆库中抗体加入,形成新种群
% individuals input 抗体群
% sizepop input 抗体数
% bestindividuals input 记忆库
% overbest input 记忆库容量
m = sizepop+overbest;
newindividuals = struct('fitness',zeros(1,m), 'concentration',zeros(1,m),'excellence',zeros(1,m),'chrom',[]);
% 遗传操作得到的抗体
for i=1:sizepop
newindividuals.fitness(i) = individuals.fitness(i);
newindividuals.concentration(i) = individuals.concentration(i);
newindividuals.excellence(i) = individuals.excellence(i);
newindividuals.chrom(i,:) = individuals.chrom(i,:);
end
% 记忆库中抗体
for i=sizepop+1:m
newindividuals.fitness(i) = bestindividuals.fitness(i-sizepop);
newindividuals.concentration(i) = bestindividuals.concentration(i-sizepop);
newindividuals.excellence(i) = bestindividuals.excellence(i-sizepop);
newindividuals.chrom(i,:) = bestindividuals.chrom(i-sizepop,:);
end
end
function psd=popinit(M,length)
ss=[];
for i=1:M
a=randperm(31,length);
ss(i,:)=a;
end
psd=ss;
end
function rets=bestselect(individuals,m,n)
% 初始化记忆库,依据excellence,将群体中高适应度低相似度的overbest个个体存入记忆库
% m input 抗体数
% n input 记忆库个体数\父代群规模
% individuals input 抗体群
% bestindividuals output 记忆库\父代群
% 精英保留策略,将fitness最好的s个个体先存起来,避免因其浓度高而被淘汰
s=3;
rets=struct('fitness',zeros(1,n), 'concentration',zeros(1,n),'excellence',zeros(1,n),'chrom',[]);
[fitness,index] = sort(individuals.fitness);
for i=1:s
rets.fitness(i) = individuals.fitness(index(i));
rets.concentration(i) = individuals.concentration(index(i));
rets.excellence(i) = individuals.excellence(index(i));
rets.chrom(i,:) = individuals.chrom(index(i),:);
end
% 剩余m-s个个体
leftindividuals=struct('fitness',zeros(1,m-s), 'concentration',zeros(1,m-s),'excellence',zeros(1,m-s),'chrom',[]);
for k=1:m-s
leftindividuals.fitness(k) = individuals.fitness(index(k+s));
leftindividuals.concentration(k) = individuals.concentration(index(k+s));
leftindividuals.excellence(k) = individuals.excellence(index(k+s));
leftindividuals.chrom(k,:) = individuals.chrom(index(k+s),:);
end
% 将剩余抗体按excellence值排序
[excellence,index]=sort(1./leftindividuals.excellence);
% 在剩余抗体群中按excellence再选n-s个最好的个体
for i=s+1:n
rets.fitness(i) = leftindividuals.fitness(index(i-s));
rets.concentration(i) = leftindividuals.concentration(index(i-s));
rets.excellence(i) = leftindividuals.excellence(index(i-s));
rets.chrom(i,:) = leftindividuals.chrom(index(i-s),:);
end
end
function ret=Cross(pcross,chrom,sizepop,length)
% 交叉操作
% pcorss input : 交叉概率
% chrom input : 抗体群
% sizepop input : 种群规模
% length input : 抗体长度
% ret output : 交叉得到的抗体群
% 每一轮for循环中,可能会进行一次交叉操作,随机选择染色体是和交叉位置,是否进行交叉操作则由交叉概率(continue)控制
for i=1:sizepop
% 随机选择两个染色体进行交叉
pick=rand;
while prod(pick)==0
pick=rand(1);
end
if pick>pcross
continue;
end
% 找出交叉个体
index(1)=unidrnd(sizepop);
index(2)=unidrnd(sizepop);
while index(2)==index(1)
index(2)=unidrnd(sizepop);
end
% 选择交叉位置
pos=ceil(length*rand);
while pos==1
pos=ceil(length*rand);
end
% 个体交叉
chrom1=chrom(index(1),:);
chrom2=chrom(index(2),:);
k=chrom1(pos:length);
chrom1(pos:length)=chrom2(pos:length);
chrom2(pos:length)=k;
% 满足约束条件赋予新种群
flag1=test(chrom(index(1),:));
flag2=test(chrom(index(2),:));
if flag1*flag2==1
chrom(index(1),:)=chrom1;
chrom(index(2),:)=chrom2;
end
end
ret=chrom;
end
function flag=test(code)
% 检查个体是否满足距离约束
% code input 个体
% flag output 是否满足要求标志
city_coordinate=[1304,2312;3639,1315;4177,2244;3712,1399;3488,1535;3326,1556;3238,1229;4196,1044;4312,790;4386,570;
3007,1970;2562,1756;2788,1491;2381,1676;1332,695;3715,1678;3918,2179;4061,2370;3780,2212;3676,2578;
4029,2838;4263,2931;3429,1908;3507,2376;3394,2643;3439,3201;2935,3240;3140,3550;2545,2357;2778,2826;2370,2975];
flag=1;
if max( max(dist( city_coordinate(code,:)') ) )>3000
flag=0;
end
end
结果展示如下: