欢迎关注我的CSDN:https://spike.blog.csdn.net/
本文地址:https://blog.csdn.net/caroline_wendy/article/details/131423024
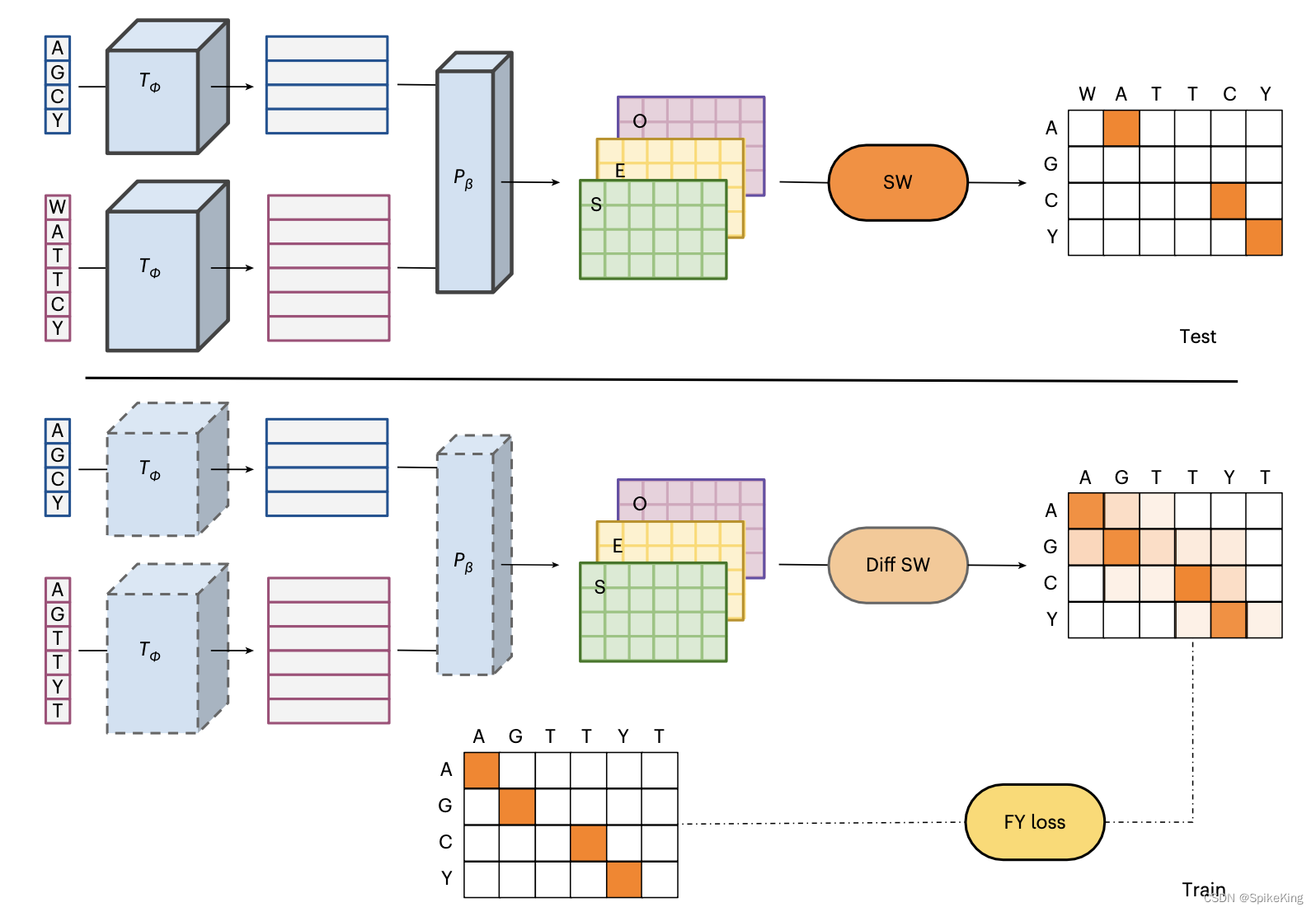
DEDAL:Deep Embedding and Differentiable ALignment
这篇论文提出一种新的蛋白质序列比对算法,叫做 DEDAL (深度嵌入和可微分对齐)。利用最新的深度学习语言模型和可微分编程的技术,学习一个能够根据输入序列自适应地生成替换得分和缺口惩罚的模型。在大量的原始蛋白质序列和已知正确对齐的序列对上进行训练,然后用标准的 Smith–Waterman (史密斯-沃特曼) 算法来寻找最优的局部对齐。实验结果表明,DEDAL在远缘同源序列上的对齐准确度,和同源检测能力都显著优于现有的方法,尤其是在低序列相似度的情况下。
这篇论文的创新点主要有以下几个方面:
- 提出了一种灵活的参数化方法,使得替换得分和缺口惩罚能够根据每对序列和每对位置的上下文信息进行调整,而不是使用固定的矩阵和参数。
- 使用了一个基于 Transformer 的深度语言模型,将离散的蛋白质序列映射到一个连续的向量空间,从而能够捕捉序列之间的复杂相似性。
- 设计了一个可微分的 Smith–Waterman 算法,使得模型可以通过端到端的梯度优化来最小化对齐误差,并且可以同时利用大量的无标注序列和有标注序列进行训练。
使用 DEDAL 计算序列的同源性,作为 MSA 中 seq 筛选的依据。
相关信息:
-
GitHub:https://github.com/google-research/google-research/tree/master/dedal
下载工程:
vim ~/.ssh/id_rsa
chmod 600 ~/.ssh/id_rsa
git clone https://github.com/google-research/google-research.git
工程较多,大约 523.91 MiB,下载较慢。
下载模型:https://tfhub.dev/google/dedal/3,大约 172 MB,上传至百度网盘,位置:DEDAL_Deep_Embedding_and_Differentiable_ALignment
1. 配置 Tensorflow 环境
构建运行环境:
conda env list
conda create -n dedal python=3.8 -y
conda activate dedal
待安装 Python 包,建议手动安装:
cd google-research/dedal
cat requirements.txt
# 待安装的包,建议手动安装:
absl-py>=0.7.0
gin-config>=0.4.0
numpy>=1.18.4
tensorflow>=2.3.0
tensorflow_datasets>=3.0.0
tensorflow_probability>=0.1.0
tf-models-nightly
安装 TensorFlow 推理框架:
nvcc --version
# cuda 11.7 也可以安装 11.3
conda install cudatoolkit=11.3.1 cudnn=8.2.1
pip install tensorflow-gpu==2.7.0
pip install protobuf==3.20.3 # 否则报错
安装 locate 命令 (安装时间较长,建议不安装):
sudo apt update && sudo apt install mlocate
卡在 Initializing mlocate database; this may take some time…
导出 libcudnn.so.8 至当前环境:
locate libcudnn.so.8 # 也可以手动搜索
vim ~/.bashrc
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/root/miniconda3/pkgs/cudnn-8.2.1-cuda11.3_0/lib/
source ~/.bashrc
测试 TensorFlow 是否安装成功:
python3
import tensorflow as tf
print(f"is_gpu_available: {
tf.test.is_gpu_available()}")
gpu_device_name = tf.test.gpu_device_name()
print(f"gpu_device_name: {
gpu_device_name}")
from tensorflow.python.client import device_lib
# 列出所有的本地机器设备
local_device_protos = device_lib.list_local_devices()
# 只打印GPU设备
print(x) for x in local_device_protos if x.device_type == 'GPU'
验证成功之后,再安装其他的包,包括tensorflow_datasets、tensorflow_probability、tf-models-official:
pip install tensorflow_datasets==4.9.2 tensorflow_probability==0.20.1 tf-models-official==2.12.0
建议使用 tf-models-official,而不是 tf-models-nightly。
2. 使用 DEDAL 模型
使用 bypy 下载模型,或已下载的模型:
mkdir mydata
cd mydata/
bypy downfile DEDAL_Deep_Embedding_and_Differentiable_ALignment/dedal_3.tar.gz dedal_3.tar.gz
tar zxvf dedal_3.tar.gz
已经添加日志的测试脚本,注意替换hub.load的加载路径,如下:
import os
os.environ["TF_CPP_MIN_LOG_LEVEL"] = "2" # 避免 TF 的垃圾日志
os.environ["CUDA_VISIBLE_DEVICES"] = "1"
os.environ["TF_FORCE_GPU_ALLOW_GROWTH"] = "true"
import tensorflow as tf
import tensorflow_hub as hub
from dedal import infer # Requires google_research/google-research.
gpus = tf.config.experimental.list_physical_devices(device_type='GPU')
for gpu in gpus:
tf.config.experimental.set_memory_growth(gpu, True)
# tf.config.experimental.set_visible_devices(gpus[1], device_type='GPU')
# tf.config.experimental.set_memory_growth(1, True)
print("[Info] GPU is ready!")
# "Gorilla" and "Mallard" sequences from [1, Figure 3].
protein_a = 'SVCCRDYVRYRLPLRVVKHFYWTSDSCPRPGVVLLTFRDKEICADPRVPWVKMILNKL'
protein_b = 'VKCKCSRKGPKIRFSNVRKLEIKPRYPFCVEEMIIVTLWTRVRGEQQHCLNPKRQNTVRLLKWY'
# Represents sequences as `tf.Tensor<tf.float32>[2, 512]` batch of tokens.
dedal_model = hub.load('dedal/mydata/') # 耗时较长
print("[Info] Model is ready!")
inputs = infer.preprocess(protein_a, protein_b)
print(f"[Info] infer preprocess is pass! device: {
inputs.device}")
# Aligns `protein_a` to `protein_b`.
align_out = dedal_model(inputs) # 耗时较长,字典类型,无法使用device
print(f"[Info] dedal_model is pass!")
# Retrieves per-position embeddings of both sequences.
embeddings = dedal_model.call(inputs, embeddings_only=True)
print(f"[Info] dedal_model embeddings is pass! device: {
embeddings.device}")
# Postprocesses output and displays alignment.
output = infer.expand([align_out['sw_scores'], align_out['paths'], align_out['sw_params']])
print(f"[Info] infer expand is pass!")
output = infer.postprocess(output, len(protein_a), len(protein_b))
print(f"[Info] infer postprocess is pass!")
alignment = infer.Alignment(protein_a, protein_b, *output)
print(f"[Info] infer Alignment is pass!")
print(alignment)
# Displays the raw Smith-Waterman score and the homology detection logits.
print('Smith-Waterman score (uncorrected):', align_out['sw_scores'].numpy(), align_out['sw_scores'].device)
print('Homology detection logits:', align_out['homology_logits'].numpy(), align_out['homology_logits'].device)
输入日志如下:
[Info] GPU is ready!
[Info] Model is ready!
[Info] infer preprocess is pass! device: /job:localhost/replica:0/task:0/device:GPU:0
[Info] dedal_model is pass!
[Info] dedal_model embeddings is pass! device: /job:localhost/replica:0/task:0/device:GPU:0
[Info] infer expand is pass!
[Info] infer postprocess is pass!
[Info] infer Alignment is pass!
0 SVC-CRDYVRYRLPLRVVKHFYW--TSDSCPRPGVV-LLTF---RDKEICADPRVPWVKMILNKL 57
::| |.:.: ..:...:|::::. .:::|:.:.:: .|.. ..:::|::|:::.::::|:::
0 VKCKCSRKG-PKIRFSNVRKLEIKPRYPFCVEEMIIVTLWTRVRGEQQHCLNPKRQNTVRLLKWY 63
Smith-Waterman score (uncorrected): [14.423605] /job:localhost/replica:0/task:0/device:GPU:0
Homology detection logits: [11.416809] /job:localhost/replica:0/task:0/device:GPU:0
注意,DEDAL 项目并不依赖于 Google Research 工程,支持直接复制单独运行。
Bugfix
Bug1: If this call came from a _pb2.py file, your generated code is out of date and must be regenerated with protoc >= 3.19.0.
原因是 protobuf 的版本不兼容,建议降低版本即可:
pip install protobuf== # 不指定版本号,查看版本
pip install protobuf==3.20.3 # 更新版本,否则报错
Bug2: tensorflow/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library ‘libcudnn.so.8’
导出 libcudnn.so.8 至当前环境:
locate libcudnn.so.8
vim /etc/profile
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/[your path]/miniconda3/pkgs/cudnn-8.2.1-cuda11.3_0/lib/
source /etc/profile
Bug3: oneDNN custom operations are on
完整日志如下:
2023-06-27 07:23:36.909581: I tensorflow/core/util/port.cc:110] oneDNN custom operations are on. You may see slightly different numerical results due to floating-point round-off errors from different computation orders. To turn them off, set the environment variable `TF_ENABLE_ONEDNN_OPTS=0`.
输出:
export TF_ENABLE_ONEDNN_OPTS=0
参考
- CSDN - TensorFlow GPU 与 源码编译
- conda查找安装包的版本以及安装特定版本的包
- CUDA 11.7 版本下安装Tensorflow-gpu以及Keras (Windows以及Ubuntu)
- 已解决If this call came from a _pb2.py file, your generated code is out of date and must be regenerated
- Linux 安装locate 命令
- tensorflow如何使用gpu
- StackOverflow - tensorflow store training data on GPU memory
- Tensorflow - 使用 GPU
- GitHub - You must feed a value for placeholder tensor … with dtype int32
- StackOverflow - How can I know whether a tensorflow tensor is in cuda or cpu?