一:数据集介绍
1:数据集下载
https://archive.ics.uci.edu/ml/datasets/Wine+Quality

我这里选择的是红酒样本
数据的特征与标签
特征:11个 ; 标签:红酒质量0-10之间,11个类别

2:查看数据集
可以看到数据都在一列里,需要改一下
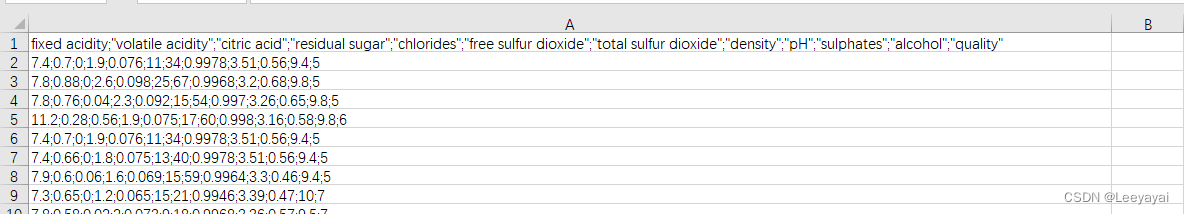
二:数据处理
1:数据分列
观察数据,在一列里用分号隔开,由此对数据分列
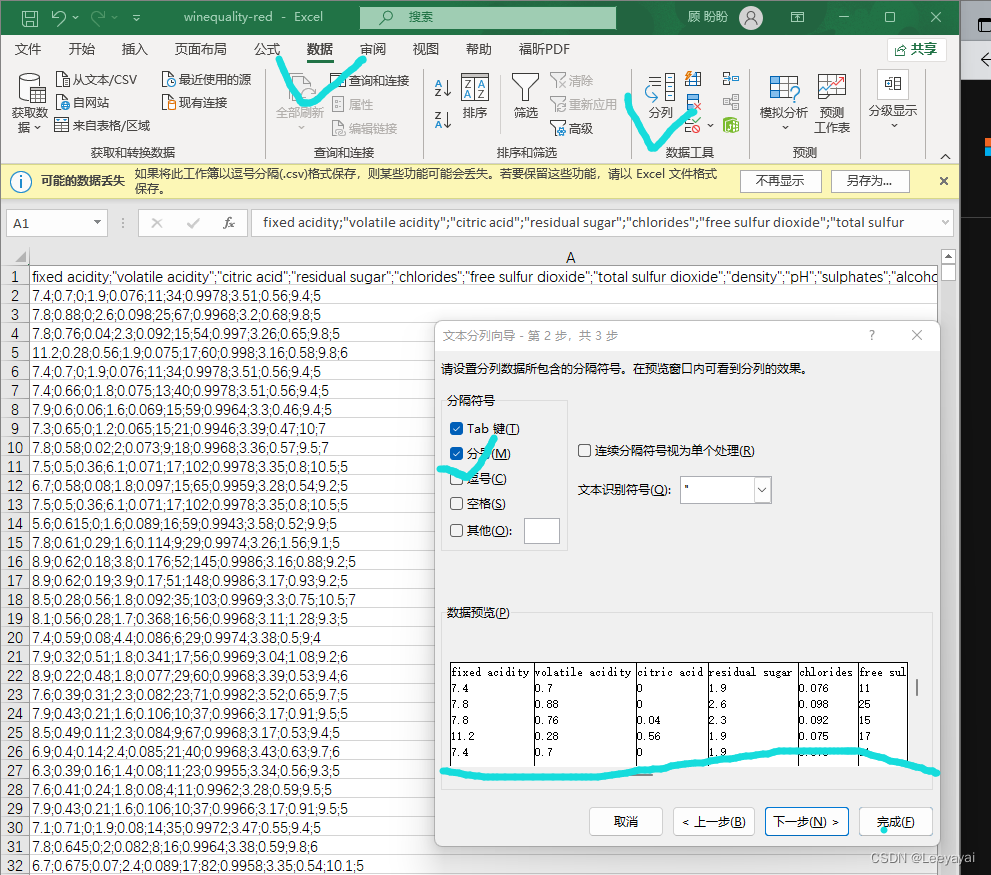
选定需要分列的数据–选数据菜单–分列–分隔符–选分号–OK
分列后的数据

2:导入数据
import pandas as pd
#获取数据
data = pd.read_csv("F:\\书籍学习:python数据挖掘与机器学习实战\\葡萄酒数据集的随机森林分类\\winequality-red.csv")
data.head()#查看数据
| fixed acidity | volatile acidity | citric acid | residual sugar | chlorides | free sulfur dioxide | total sulfur dioxide | density | pH | sulphates | alcohol | quality | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 7.4 | 0.70 | 0.00 | 1.9 | 0.076 | 11.0 | 34.0 | 0.9978 | 3.51 | 0.56 | 9.4 | 5 |
| 1 | 7.8 | 0.88 | 0.00 | 2.6 | 0.098 | 25.0 | 67.0 | 0.9968 | 3.20 | 0.68 | 9.8 | 5 |
| 2 | 7.8 | 0.76 | 0.04 | 2.3 | 0.092 | 15.0 | 54.0 | 0.9970 | 3.26 | 0.65 | 9.8 | 5 |
| 3 | 11.2 | 0.28 | 0.56 | 1.9 | 0.075 | 17.0 | 60.0 | 0.9980 | 3.16 | 0.58 | 9.8 | 6 |
| 4 | 7.4 | 0.70 | 0.00 | 1.9 | 0.076 | 11.0 | 34.0 | 0.9978 | 3.51 | 0.56 | 9.4 | 5 |
# 导入所有需要的库
import numpy as np
import pandas as pd
from sklearn.ensemble import RandomForestClassifier
3:将数据拆分为特征与标签
features = data.drop('quality', 1)
# df = data.iloc[:, :11] #取前11列数据
labels = data['quality']
print(features.shape)
print(labels.shape)
(1599, 11)
(1599,)
C:\Users\Hp\AppData\Local\Temp\ipykernel_12320\351942566.py:1: FutureWarning: In a future version of pandas all arguments of DataFrame.drop except for the argument 'labels' will be keyword-only.
features = data.drop('quality', 1)
三:数据分析
1:数据的描述性分析
# 描述性分析
print(features.describe())
# 直方图
# hist(),输出各个特征对比的直方图
features.hist()
fixed acidity volatile acidity citric acid residual sugar \
count 1599.000000 1599.000000 1599.000000 1599.000000
mean 8.319637 0.527821 0.270976 2.538806
std 1.741096 0.179060 0.194801 1.409928
min 4.600000 0.120000 0.000000 0.900000
25% 7.100000 0.390000 0.090000 1.900000
50% 7.900000 0.520000 0.260000 2.200000
75% 9.200000 0.640000 0.420000 2.600000
max 15.900000 1.580000 1.000000 15.500000
chlorides free sulfur dioxide total sulfur dioxide density \
count 1599.000000 1599.000000 1599.000000 1599.000000
mean 0.087467 15.874922 46.467792 0.996747
std 0.047065 10.460157 32.895324 0.001887
min 0.012000 1.000000 6.000000 0.990070
25% 0.070000 7.000000 22.000000 0.995600
50% 0.079000 14.000000 38.000000 0.996750
75% 0.090000 21.000000 62.000000 0.997835
max 0.611000 72.000000 289.000000 1.003690
pH sulphates alcohol
count 1599.000000 1599.000000 1599.000000
mean 3.311113 0.658149 10.422983
std 0.154386 0.169507 1.065668
min 2.740000 0.330000 8.400000
25% 3.210000 0.550000 9.500000
50% 3.310000 0.620000 10.200000
75% 3.400000 0.730000 11.100000
max 4.010000 2.000000 14.900000
array([[<AxesSubplot:title={'center':'fixed acidity'}>,
<AxesSubplot:title={'center':'volatile acidity'}>,
<AxesSubplot:title={'center':'citric acid'}>],
[<AxesSubplot:title={'center':'residual sugar'}>,
<AxesSubplot:title={'center':'chlorides'}>,
<AxesSubplot:title={'center':'free sulfur dioxide'}>],
[<AxesSubplot:title={'center':'total sulfur dioxide'}>,
<AxesSubplot:title={'center':'density'}>,
<AxesSubplot:title={'center':'pH'}>],
[<AxesSubplot:title={'center':'sulphates'}>,
<AxesSubplot:title={'center':'alcohol'}>, <AxesSubplot:>]],
dtype=object)

2:各等级酒的描述性分析
分为三个等级:低级(0-3),中级(4-7),高级(8-10)
(最大最小值,平均值,标准差)
2.1: 统计表类别个数df.value_counts()
#查看标签值,有几类标签
print(labels.value_counts())
5 681
6 638
7 199
4 53
8 18
3 10
Name: quality, dtype: int64
2.2:对数据进行分割,低级,中级,高级红酒
暂放-
3 :变量的相关性分析
1:np.corrcoef()

#features = data.drop('quality', 1)
df = data.iloc[:, :11] #取前11列数据
#print(df.head())#查看前5列数据
#分析两个变量间的相关性
print(np.corrcoef(data.iloc[1], data.iloc[2]))
#分析所有变量之间的相关性
print(np.corrcoef(df, rowvar = False))
[[1. 0.99368451]
[0.99368451 1. ]]
[[ 1. -0.25613089 0.67170343 0.11477672 0.09370519 -0.15379419
-0.11318144 0.66804729 -0.68297819 0.18300566 -0.06166827]
[-0.25613089 1. -0.55249568 0.00191788 0.06129777 -0.01050383
0.07647 0.02202623 0.23493729 -0.26098669 -0.20228803]
[ 0.67170343 -0.55249568 1. 0.14357716 0.20382291 -0.06097813
0.03553302 0.36494718 -0.54190414 0.31277004 0.10990325]
[ 0.11477672 0.00191788 0.14357716 1. 0.05560954 0.187049
0.20302788 0.35528337 -0.08565242 0.00552712 0.04207544]
[ 0.09370519 0.06129777 0.20382291 0.05560954 1. 0.00556215
0.04740047 0.20063233 -0.26502613 0.37126048 -0.22114054]
[-0.15379419 -0.01050383 -0.06097813 0.187049 0.00556215 1.
0.66766645 -0.02194583 0.0703775 0.05165757 -0.06940835]
[-0.11318144 0.07647 0.03553302 0.20302788 0.04740047 0.66766645
1. 0.07126948 -0.06649456 0.04294684 -0.20565394]
[ 0.66804729 0.02202623 0.36494718 0.35528337 0.20063233 -0.02194583
0.07126948 1. -0.34169933 0.14850641 -0.49617977]
[-0.68297819 0.23493729 -0.54190414 -0.08565242 -0.26502613 0.0703775
-0.06649456 -0.34169933 1. -0.1966476 0.20563251]
[ 0.18300566 -0.26098669 0.31277004 0.00552712 0.37126048 0.05165757
0.04294684 0.14850641 -0.1966476 1. 0.09359475]
[-0.06166827 -0.20228803 0.10990325 0.04207544 -0.22114054 -0.06940835
-0.20565394 -0.49617977 0.20563251 0.09359475 1. ]]
2:pandas用法,df为datafram数据–df.corr()
print(df.corr())
fixed acidity volatile acidity citric acid \
fixed acidity 1.000000 -0.256131 0.671703
volatile acidity -0.256131 1.000000 -0.552496
citric acid 0.671703 -0.552496 1.000000
residual sugar 0.114777 0.001918 0.143577
chlorides 0.093705 0.061298 0.203823
free sulfur dioxide -0.153794 -0.010504 -0.060978
total sulfur dioxide -0.113181 0.076470 0.035533
density 0.668047 0.022026 0.364947
pH -0.682978 0.234937 -0.541904
sulphates 0.183006 -0.260987 0.312770
alcohol -0.061668 -0.202288 0.109903
residual sugar chlorides free sulfur dioxide \
fixed acidity 0.114777 0.093705 -0.153794
volatile acidity 0.001918 0.061298 -0.010504
citric acid 0.143577 0.203823 -0.060978
residual sugar 1.000000 0.055610 0.187049
chlorides 0.055610 1.000000 0.005562
free sulfur dioxide 0.187049 0.005562 1.000000
total sulfur dioxide 0.203028 0.047400 0.667666
density 0.355283 0.200632 -0.021946
pH -0.085652 -0.265026 0.070377
sulphates 0.005527 0.371260 0.051658
alcohol 0.042075 -0.221141 -0.069408
total sulfur dioxide density pH sulphates \
fixed acidity -0.113181 0.668047 -0.682978 0.183006
volatile acidity 0.076470 0.022026 0.234937 -0.260987
citric acid 0.035533 0.364947 -0.541904 0.312770
residual sugar 0.203028 0.355283 -0.085652 0.005527
chlorides 0.047400 0.200632 -0.265026 0.371260
free sulfur dioxide 0.667666 -0.021946 0.070377 0.051658
total sulfur dioxide 1.000000 0.071269 -0.066495 0.042947
density 0.071269 1.000000 -0.341699 0.148506
pH -0.066495 -0.341699 1.000000 -0.196648
sulphates 0.042947 0.148506 -0.196648 1.000000
alcohol -0.205654 -0.496180 0.205633 0.093595
alcohol
fixed acidity -0.061668
volatile acidity -0.202288
citric acid 0.109903
residual sugar 0.042075
chlorides -0.221141
free sulfur dioxide -0.069408
total sulfur dioxide -0.205654
density -0.496180
pH 0.205633
sulphates 0.093595
alcohol 1.000000
3:绘图
3.1:散点图–seaborn或者pandas
此处只取前3列数据
第一行代码结果如图所示,是一张大图,其中包含9个子图,每个子图都是每个维度和其他某个维度的相关关系图,这其中主对角线上的图,则是每个维度的数据分布直方图。
而第二行代码是画出同样的图形,但却以fixed acidity(第一列数据)这个维度的数据为标准,从图中可以看出,sepal_width这列数据共5个不同的数值,每个数值一种颜色,所以生成的图是彩色的。
import scipy.stats as ss
import seaborn as sns ##导入库
dff=data.iloc[:, :3]
sns.pairplot(dff)
sns.pairplot(dff , hue ='fixed acidity')
<seaborn.axisgrid.PairGrid at 0x1a1d70fda30>


3.2:热力图–heatmap()
import scipy.stats as ss
import seaborn as sns ##导入库
import matplotlib.pyplot as plt
figure, ax = plt.subplots(figsize=(12, 12))
sns.heatmap(dff.corr(), square=True, annot=True, ax=ax)
<AxesSubplot:>

这个颜色太丑了–换一个
博文详解
https://blog.csdn.net/weixin_45492560/article/details/106227864
颜色参数:
cmap:指定一个colormap对象,用于热力图的填充色
center:指定颜色中心值,通过该参数可以调整热力图的颜色深浅

figure, ax = plt.subplots(figsize=(12, 12))
sns.heatmap(dff.corr(),cmap='GnBu' ,square=True, annot=True, ax=ax)
<AxesSubplot:>

figure, ax = plt.subplots(figsize=(12, 12))
sns.heatmap(dff.corr(),cmap='YlGnBu' ,square=True, annot=True, ax=ax)
<AxesSubplot:>

figure, ax = plt.subplots(figsize=(12, 12))
sns.heatmap(dff.corr(),cmap='summer' ,square=True, annot=True, ax=ax)
<AxesSubplot:>

四:使用随机森林构建模型
1:使用模型前的数据处理

# 特征与标签
features = data.drop('quality', 1)
# df = data.iloc[:, :11] #取前11列数据
labels = data['quality']
print(features.shape)
print(labels.shape)
# 拆分训练集与测试集
# 构造训练集和测试集
# <pre name="code" class="python"><span style="font-size:14px;">
from sklearn.model_selection import train_test_split
# 交叉验证
X_train,X_test,y_train,y_test=train_test_split(features,labels,random_state=1,test_size=0.3)
# print(X_train.shape)
# print(X_test.shape)
# print(y_train.shape)
# print(y_test.shape)
# 默认为75%为训练,25%为测试
(1599, 11)
(1599,)
C:\Users\Hp\AppData\Local\Temp\ipykernel_12320\1883349980.py:2: FutureWarning: In a future version of pandas all arguments of DataFrame.drop except for the argument 'labels' will be keyword-only.
features = data.drop('quality', 1)
2:复习sklearn建模的基本流程
3: 建模与分析
画出随机森林和决策树在一组交叉验证下的效果对比
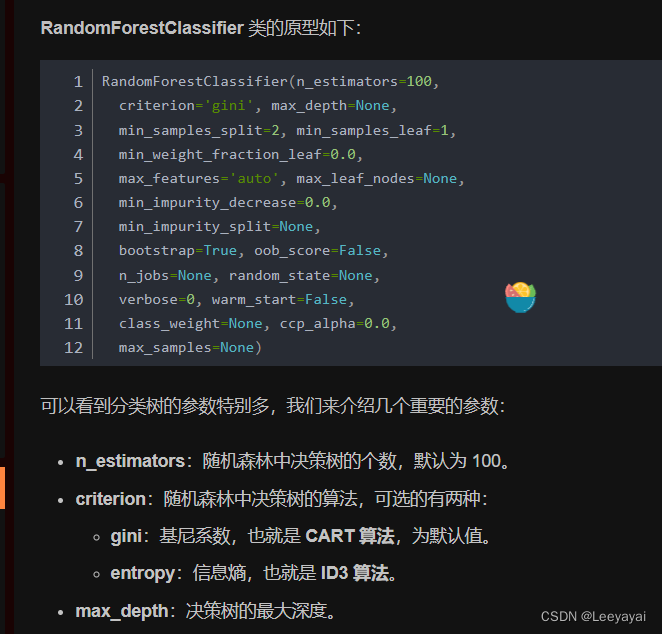
# 使用默认参数
model = RandomForestClassifier(oob_score=True, random_state=10)
model.fit(X_train,y_train)
test_predict = model.predict(X_test)
from sklearn.metrics import accuracy_score
accuracy_score(y_test, test_predict)
0.6979166666666666
from sklearn.model_selection import cross_val_score
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
import matplotlib.pyplot as plt
rfc = RandomForestClassifier(n_estimators=25)
rfc_s = cross_val_score(rfc,X_train,y_train,cv=10)
# 交叉验证划分为10折,
clf = DecisionTreeClassifier()
clf_s = cross_val_score(clf,X_train,y_train,cv=10)
plt.plot(range(1,11),rfc_s,label = "RandomForest")
plt.plot(range(1,11),clf_s,label = "Decision Tree")
plt.legend()
plt.show()
D:\anaconda\lib\site-packages\sklearn\model_selection\_split.py:676: UserWarning: The least populated class in y has only 8 members, which is less than n_splits=10.
warnings.warn(
D:\anaconda\lib\site-packages\sklearn\model_selection\_split.py:676: UserWarning: The least populated class in y has only 8 members, which is less than n_splits=10.
warnings.warn(

画出随机森林和决策树在10组交叉验证下的效果比较
rfc_l = []
clf_l = []
for i in range(10):
rfc = RandomForestClassifier(n_estimators=25)
rfc_s = cross_val_score(rfc,X_train,y_train,cv=10).mean()
rfc_l.append(rfc_s)
clf = DecisionTreeClassifier()
clf_s = cross_val_score(clf,X_train,y_train,cv=10).mean()
clf_l.append(clf_s)
plt.plot(range(1,11),rfc_l,label = "Random Forest")
plt.plot(range(1,11),clf_l,label = "Decision Tree")
plt.legend()
plt.show()
#是否有注意到,单个决策树的波动轨迹和随机森林一致?
#再次验证了我们之前提到的,单个决策树的准确率越高,随机森林的准确率也会越高
D:\anaconda\lib\site-packages\sklearn\model_selection\_split.py:676: UserWarning: The least populated class in y has only 8 members, which is less than n_splits=10.
warnings.warn(
D:\anaconda\lib\site-packages\sklearn\model_selection\_split.py:676: UserWarning: The least populated class in y has only 8 members, which is less than n_splits=10.
warnings.warn(
D:\anaconda\lib\site-packages\sklearn\model_selection\_split.py:676: UserWarning: The least populated class in y has only 8 members, which is less than n_splits=10.
warnings.warn(
D:\anaconda\lib\site-packages\sklearn\model_selection\_split.py:676: UserWarning: The least populated class in y has only 8 members, which is less than n_splits=10.
warnings.warn(
D:\anaconda\lib\site-packages\sklearn\model_selection\_split.py:676: UserWarning: The least populated class in y has only 8 members, which is less than n_splits=10.
warnings.warn(
D:\anaconda\lib\site-packages\sklearn\model_selection\_split.py:676: UserWarning: The least populated class in y has only 8 members, which is less than n_splits=10.
warnings.warn(
D:\anaconda\lib\site-packages\sklearn\model_selection\_split.py:676: UserWarning: The least populated class in y has only 8 members, which is less than n_splits=10.
warnings.warn(
D:\anaconda\lib\site-packages\sklearn\model_selection\_split.py:676: UserWarning: The least populated class in y has only 8 members, which is less than n_splits=10.
warnings.warn(
D:\anaconda\lib\site-packages\sklearn\model_selection\_split.py:676: UserWarning: The least populated class in y has only 8 members, which is less than n_splits=10.
warnings.warn(
D:\anaconda\lib\site-packages\sklearn\model_selection\_split.py:676: UserWarning: The least populated class in y has only 8 members, which is less than n_splits=10.
warnings.warn(
D:\anaconda\lib\site-packages\sklearn\model_selection\_split.py:676: UserWarning: The least populated class in y has only 8 members, which is less than n_splits=10.
warnings.warn(
D:\anaconda\lib\site-packages\sklearn\model_selection\_split.py:676: UserWarning: The least populated class in y has only 8 members, which is less than n_splits=10.
warnings.warn(
D:\anaconda\lib\site-packages\sklearn\model_selection\_split.py:676: UserWarning: The least populated class in y has only 8 members, which is less than n_splits=10.
warnings.warn(
D:\anaconda\lib\site-packages\sklearn\model_selection\_split.py:676: UserWarning: The least populated class in y has only 8 members, which is less than n_splits=10.
warnings.warn(
D:\anaconda\lib\site-packages\sklearn\model_selection\_split.py:676: UserWarning: The least populated class in y has only 8 members, which is less than n_splits=10.
warnings.warn(
D:\anaconda\lib\site-packages\sklearn\model_selection\_split.py:676: UserWarning: The least populated class in y has only 8 members, which is less than n_splits=10.
warnings.warn(
D:\anaconda\lib\site-packages\sklearn\model_selection\_split.py:676: UserWarning: The least populated class in y has only 8 members, which is less than n_splits=10.
warnings.warn(
D:\anaconda\lib\site-packages\sklearn\model_selection\_split.py:676: UserWarning: The least populated class in y has only 8 members, which is less than n_splits=10.
warnings.warn(
D:\anaconda\lib\site-packages\sklearn\model_selection\_split.py:676: UserWarning: The least populated class in y has only 8 members, which is less than n_splits=10.
warnings.warn(
D:\anaconda\lib\site-packages\sklearn\model_selection\_split.py:676: UserWarning: The least populated class in y has only 8 members, which is less than n_splits=10.
warnings.warn(
