A, UNET
UNET can be called the cornerstone of medical image semantic segmentation, and often as a baseline comparison with other models. General organ segmentation, often as long as the positive normal boundaries more obvious, nothing special circumstances, the general UNET will be able to do it well.
UNET is structured as follows:

UNET Features:
FCN 1.UNET structure and very similar, but a great distinction between the two is place, the jumper (skip connection) is used at UNET connection mode, i.e. using the UNET characterized in channel dimension spliced together, form a thicker Characteristics. And a corresponding point used for fusion FCN addition , does not form a thicker feature
2.UNET network is a concept, which can change the parameters. The input image will generally UNET downsampling after 5, so that each reduced resolution general, this phase is called phase encoder. Then there are 5 times through the sample (or deconvolution using interpolation), characterized in that the resolution of FIG doubled, this stage is called a decoder stage. The intermediate stage is important to skip connection, I tried to remove or add stage skip connection layer, then the convolution, UNET effect becomes very poor.
Two, UNET ++
About UNET ++, I think UNET ++ as has been introduced in the know almost very clear: https://zhuanlan.zhihu.com/p/44958351
UNET ++ is structured as follows:

Three, Attention-UNET
Attention-UNET the following structure:
Before wherein each encoder and decoder on the resolution of the corresponding features stitching, using a AGs, re-adjust the output characteristic of the encoder. The module generates a gating signal for the importance of the different spatial characteristics of the position control, the red circle as shown in FIG.

Attention Gate (AGs) has the following structure:
AGs由有2个输入,如下图所示,分别是g和xl。g是较低分辨率的输入,如上图的绿色箭头。g个xl进入AGs前,g会先被上采样到跟xl分辨率相同。然后g和xl都会经过卷积核为1x1步长为1的卷积操作。1x1的卷积首先有降维的作用(降低通道维度),然后能让通道特征跨维度融合。经过激活函数ReLu增加非线性性,ReLu后的1x1卷积后主要作用是降维,把通道数降为1,然后激活函数sigmoid负责把特征图里的值映射到0~1之间,这时就得到注意力特征图了,最后一步就是把这个注意力特征与原来的xl相乘。得到最后的特征图。综合而言就是使用拥有较高语义特征的特征图来指导当前特征图进行注意力的选择。

四,R2U-NET
在R2U-NET论文中,此网络被用于眼球血管分割和皮肤癌分割,肺部分割任务中。R2U-NET全称为基于UNET的循环残差神经网络。
R2U-NET的结构如下:

上图中,有圈的地方就是R2U-NET中的循环结构,R2U-NET就是在UNET中嵌入了这种循环结构的网络,该循环结构详细说明如下图:
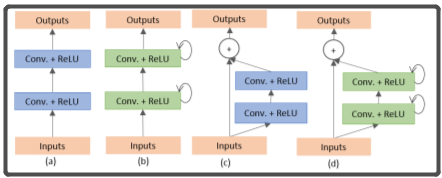
(a)为普通的两个conv模块,(b)为使用了循环卷积的模块,(c)为使用了残差卷积的模块,(d)是同时使用了残差卷积和循环卷积的模块
一个循环卷积模块其实就是t(t是可以自己设置的超参数)个普通的conv(conv+BN+ReLU)的堆叠,只不过,除了第一个conv的输入是x(输入图片)之外,后边的conv的输入是前一个conv的输出加上x作为输入。
五,CENET
CENET的结构如下:
CENET的编码器用的是resnet34。中间部分则由作者提供的DAC和RMP模块组成,DAC和RMP模块等下会有介绍。

下图中,左边的是DAC模块。可以看出作者的DAC模块很大程度地参考了inception模块,通过提供不同尺度的卷积核来获得多尺度的空间信息。感受野大的卷积操作可以为大目标提取和生成更抽象的特征,而小感受野的卷积对于小目标则更好。而右边的则是RMP模块,RMP模块由于是通过连接不同池化操作生产的特征图,由此RMP并无额外增加参数,在作者的文章中说,RMP主要用于获取上下文信息,但是我觉得这作用不打,因为医学图像的上下文信息个人认为不是十分的重要,而我在DRIVE数据集中,把RMP去掉确实影响不是十分大。


代码:
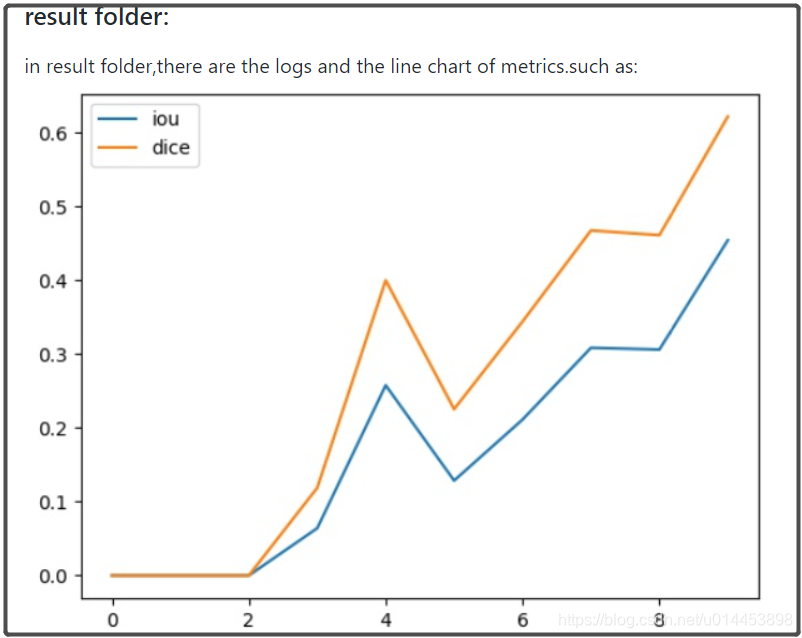
本人通过整合以上5个模型代码+FCN和SegNet,而且有7个数据集的链接,整合了一套用于基于UNET及其变体的医学图像代码。其中包括了指标可视化功能和预测结构可视化功能: