
Table of contents
Mission overview
Encoder-Decoder
skip connection
Implementation details
loss function
upsampling method
Fill or not?
How U-Net works
Mission overview
U-Net was developed for semantic segmentation tasks. When a neural network accepts an image as input, we can choose to classify objects generally or by instance. We can predict the objects contained in an image (image classification), the location of all objects (image localization/semantic segmentation), or the location of individual objects (object detection/instance segmentation).
The figure below shows the differences between these computer vision tasks. To simplify the problem, we only consider classification with one category and one label.

In a classification task, we output a vector of size k, where k is the number of categories. In the detection task, we need to output the vectors x, y, height, width, and category that define the bounding box.
But in segmentation task, we need to output an image with the same dimensions as the original input. This represents a considerable engineering challenge: how do neural networks extract relevant features from input images and then project them into segmentation masks?
Encoder-Decoder
If you are not familiar with encoder-decoder, I suggest you read this article:
https://towardsdatascience.com/understanding-latent-space-in-machine-learning-de5a7c687d8d
Encoder-decoders are related because they produce an output similar to what we want: an output with the same dimensions as the input. Can we apply the encoder-decoder concept to image segmentation? We can generate a 1D binary mask and use cross-entropy loss to train the network.
Our network consists of two parts: the encoder extracts relevant features from the image, and the decoder part takes the extracted features and reconstructs the segmentation mask.
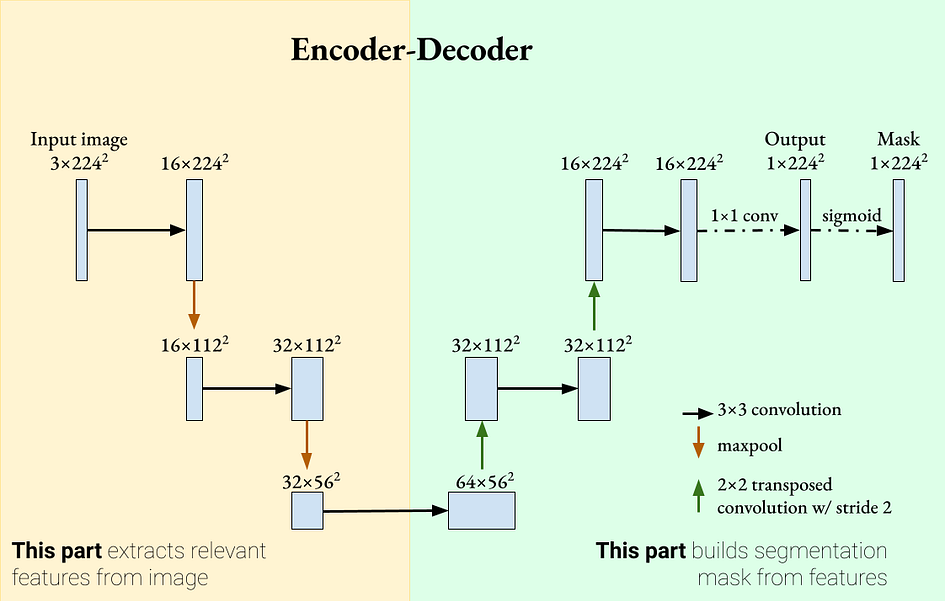
In the encoder part, convolutional layers are used, followed by ReLU and max pooling as feature extractors. In the decoder part, transposed convolution is used to increase the size of the feature map and reduce the number of channels. Padding is used to keep the size of the feature maps the same after the convolution operation.
One thing you may notice is that unlike the classification network, this network does not have fully connected/linear layers. This is an example of a fully convolutional network (FCN). FCN has been shown to perform well on segmentation tasks, starting with the paper "Fully Convolutional Networks for Semantic Segmentation" by Shelhamer et al. [1].
However, there is a problem with this network. As we add more encoder and decoder layers, we actually "shrink" the feature map more and more. Therefore, the encoder may discard more detailed features in order to obtain more general features. If we deal with medical image segmentation, it may be important for each pixel to be classified as diseased/normal. How do we ensure that this encoder-decoder network accepts both general and detailed features?
skip connection
https://towardsdatascience.com/introduction-to-resnets-c0a830a288a4
Since deep neural networks may "forget" certain features when passing information through successive layers, skip connections can reintroduce these features, making learning more powerful. Skip connections were introduced in residual networks (ResNet) and showed classification improvements as well as smoother learning gradients. Inspired by this mechanism, we can add skip connections to U-Net so that each decoder contains the feature map of its corresponding encoder. This is a defining feature of U-Net.
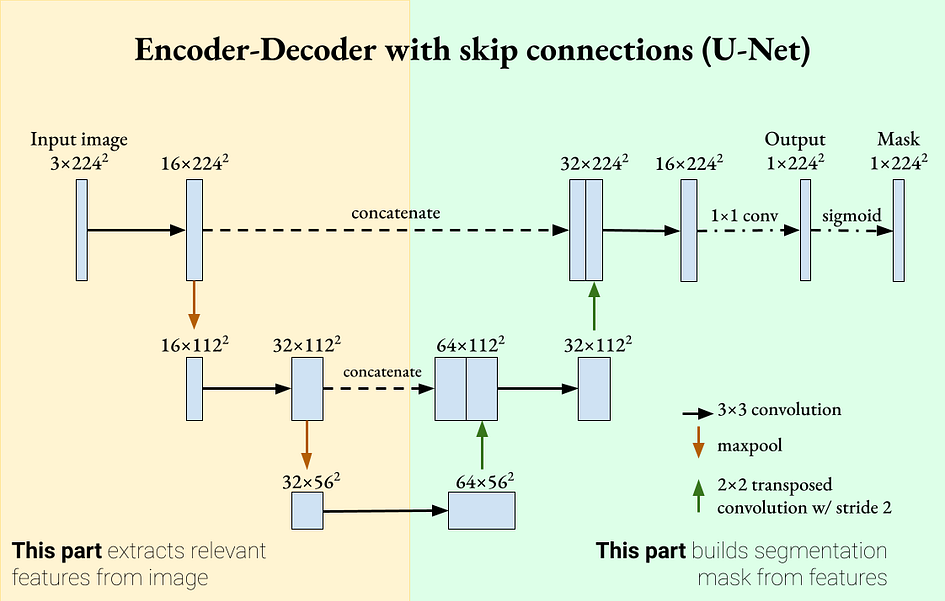
U-Net is an encoder-decoder segmentation network with skip connections. Image provided by the author. U-Net has two defining properties:
The encoder-decoder network extracts more general features as it goes deeper.
Skip connections,reintroduce detailed features in the decoder. These two properties mean that U-Net can use both detailed and general features for segmentation. U-Net was originally introduced for biomedical image processing, where segmentation accuracy is very important [2].
Implementation details

The previous sections provided a very general overview of U-Net and why it works. However, details play an important role between general understanding and practical implementation. Here I will outline some U-Net implementation choices.
loss function
Because the target is a binary mask (a pixel value of 1 means the pixel contains an object), a common loss function used to compare the output to the ground truth is a categorical cross-entropy loss (or a binary cross-entropy loss in the single-label case).
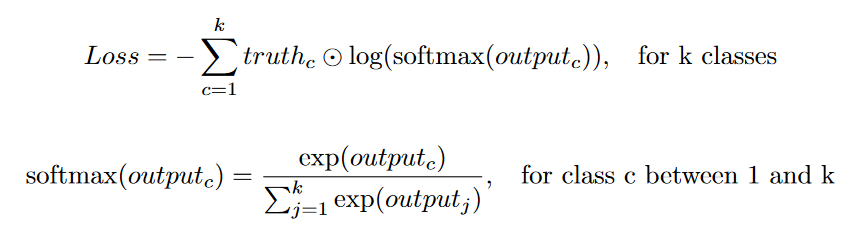
In the original U-Net paper, additional weights were added to the loss function. This weight parameter does two things: it compensates for class imbalance and gives higher importance to the segmentation boundaries. In many U-Net implementations I've found, this extra weighting factor is usually not used.
Another common loss function is Dice loss. Dice loss measures the similarity of two sets of images by comparing their intersection area to their total area. Note that Dice loss is not the same as Intersection over Union (IOU). They measure similar things, but with different denominators. The higher the Dice coefficient, the lower the Dice loss.
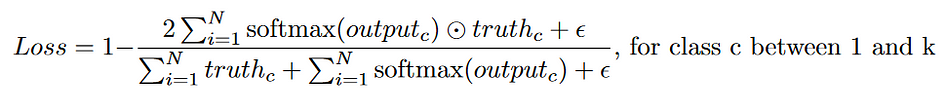
Here, an epsilon term is added to avoid dividing by 0 (epsilon is usually 1). Some implementations, such as that of Milletari et al., square the pixel values in the denominator before summing [3]. Compared to cross-entropy loss, Dice loss is very robust to imbalanced segmentation masks, which is common in biomedical image segmentation tasks.
upsampling method
Another detail is the choice of the decoder's upsampling method. Here are some common methods:
Bilinear interpolation. This method uses linear interpolation to predict output pixels. Typically, upsampling via this method is followed by a convolutional layer.
Maximum anti-pooling. This method is the reverse operation of max pooling. It uses the indexes of the max pooling operation and fills these indexes to the maximum value. All other values are set to 0. Typically, max unpooling is followed by a convolutional layer to "smooth out" any missing values.
Deconvolution/transposed convolution. There are many blog posts about deconvolution. I recommend reading this article as a good visual guide.
https://towardsdatascience.com/types-of-convolutions-in-deep-learning-717013397f4d
Deconvolution has two steps: first adding padding around each pixel of the original image, and then applying convolution. In the original U-Net, a 2x2 transposed convolution with a stride of 2 was used to change the spatial resolution and channel depth.
Pixel rearrangement. This method is used in super-resolution networks such as SRGAN. First, we use convolution to convert the C x H x W feature map to (Cr^2) x H x W. Pixel rearrangement then "rearranges" these pixels in a mosaic to produce an output of size C x (Hr) x (Wr).
Not filled or filled?
Convolutional layers, if the kernel is larger than 1x1 and without padding, will produce an output smaller than the input. This is a problem for U-Net. Recall from the U-Net diagram in the previous section that we connected a part of the image to its decoded part. If we don't use padding, then the decoded image will have smaller spatial dimensions compared to the encoded image.
However, the original U-Net paper did not use padding. Although no reason was given, I assume it is because the authors did not want to introduce segmentation errors at the edges of the image. Instead, they performed a center crop on the encoded image before concatenation. For an image with input size 572 x 572, the output will be 388 x 388, with a loss of about 50%. If you want to run U-Net without padding, you need to run it multiple times on overlapping tiles to get a complete segmentation image.
How U-Net works
Here, we implement a very simple U-Net-like network just for segmenting ellipses. This U-Net is only 3 layers deep, uses the same padding, and binary cross-entropy loss. More complex networks can use more convolutional layers per resolution, or extend the depth as needed.
import torch
import numpy as np
import torch.nn as nn
class EncoderBlock(nn.Module):
# Consists of Conv -> ReLU -> MaxPool
def __init__(self, in_chans, out_chans, layers=2, sampling_factor=2, padding="same"):
super().__init__()
self.encoder = nn.ModuleList()
self.encoder.append(nn.Conv2d(in_chans, out_chans, 3, 1, padding=padding))
self.encoder.append(nn.ReLU())
for _ in range(layers-1):
self.encoder.append(nn.Conv2d(out_chans, out_chans, 3, 1, padding=padding))
self.encoder.append(nn.ReLU())
self.mp = nn.MaxPool2d(sampling_factor)
def forward(self, x):
for enc in self.encoder:
x = enc(x)
mp_out = self.mp(x)
return mp_out, x
class DecoderBlock(nn.Module):
# Consists of 2x2 transposed convolution -> Conv -> relu
def __init__(self, in_chans, out_chans, layers=2, skip_connection=True, sampling_factor=2, padding="same"):
super().__init__()
skip_factor = 1 if skip_connection else 2
self.decoder = nn.ModuleList()
self.tconv = nn.ConvTranspose2d(in_chans, in_chans//2, sampling_factor, sampling_factor)
self.decoder.append(nn.Conv2d(in_chans//skip_factor, out_chans, 3, 1, padding=padding))
self.decoder.append(nn.ReLU())
for _ in range(layers-1):
self.decoder.append(nn.Conv2d(out_chans, out_chans, 3, 1, padding=padding))
self.decoder.append(nn.ReLU())
self.skip_connection = skip_connection
self.padding = padding
def forward(self, x, enc_features=None):
x = self.tconv(x)
if self.skip_connection:
if self.padding != "same":
# Crop the enc_features to the same size as input
w = x.size(-1)
c = (enc_features.size(-1) - w) // 2
enc_features = enc_features[:,:,c:c+w,c:c+w]
x = torch.cat((enc_features, x), dim=1)
for dec in self.decoder:
x = dec(x)
return x
class UNet(nn.Module):
def __init__(self, nclass=1, in_chans=1, depth=5, layers=2, sampling_factor=2, skip_connection=True, padding="same"):
super().__init__()
self.encoder = nn.ModuleList()
self.decoder = nn.ModuleList()
out_chans = 64
for _ in range(depth):
self.encoder.append(EncoderBlock(in_chans, out_chans, layers, sampling_factor, padding))
in_chans, out_chans = out_chans, out_chans*2
out_chans = in_chans // 2
for _ in range(depth-1):
self.decoder.append(DecoderBlock(in_chans, out_chans, layers, skip_connection, sampling_factor, padding))
in_chans, out_chans = out_chans, out_chans//2
# Add a 1x1 convolution to produce final classes
self.logits = nn.Conv2d(in_chans, nclass, 1, 1)
def forward(self, x):
encoded = []
for enc in self.encoder:
x, enc_output = enc(x)
encoded.append(enc_output)
x = encoded.pop()
for dec in self.decoder:
enc_output = encoded.pop()
x = dec(x, enc_output)
# Return the logits
return self.logits(x)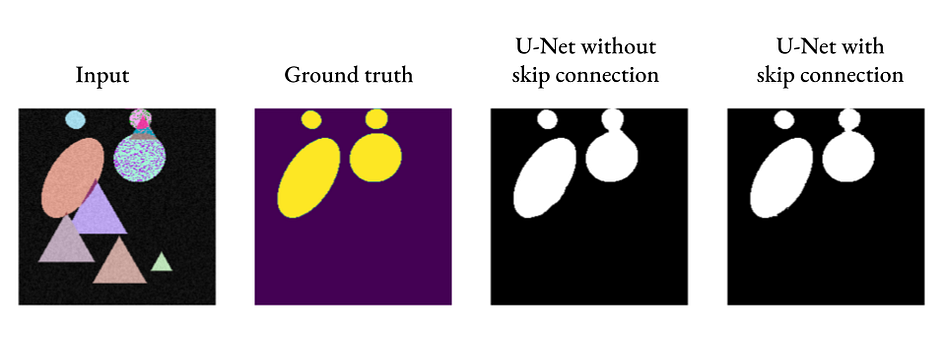
As we can see, U-Net can produce acceptable segmentation results even without skip connections, but adding skip connections can introduce finer details (see the connection between the two ovals on the right).
in conclusion
If I were to explain U-Net in one sentence, it would be that U-Net is like an encoder-decoder for images, but with skip connections to ensure that details are not lost. U-Net is frequently used in many segmentation tasks and has also achieved success in image generation tasks in recent years.
references:
[1] Long, Jonathan, Evan Shelhamer, and Trevor Darrell. “Fully convolutional networks for semantic segmentation.” Proceedings of the IEEE conference on computer vision and pattern recognition. 2015.
[2] Ronneberger, Olaf, Philipp Fischer, and Thomas Brox. “U-net: Convolutional networks for biomedical image segmentation.” International Conference on Medical image computing and computer-assisted intervention. Springer, Cham, 2015.
[3] Milletari, Fausto, Nassir Navab, and Seyed-Ahmad Ahmadi. “V-net: Fully convolutional neural networks for volumetric medical image segmentation.” 2016 fourth international conference on 3D vision (3DV). IEEE, 2016.
☆ END ☆
If you see this, it means you like this article, please forward it and like it. Search "uncle_pn" on WeChat. Welcome to add the editor's WeChat "woshicver". A high-quality blog post will be updated in the circle of friends every day.
↓ Scan the QR code to add editor↓
