The article and code have been archived in [Github warehouse: https://github.com/timerring/dive-into-AI ] or the public account [AIShareLab] can also be obtained by replying to R language .
Article directory
Sometimes data sets come from multiple places and we need to merge two or more data sets into one. The operations of merging data frames include vertical merging, horizontal merging and merging according to a common variable.
1. Vertical merge: rbind()
To merge two data frames vertically, you can use rbind( )the function. The two data frames being merged must have the same variables. This kind of merge is usually used to add observations to the data frames . For example:
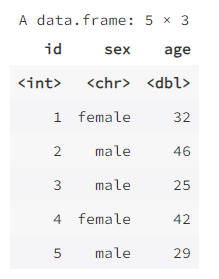
data1 <- data.frame(id = 1:5,
sex = c("female", "male", "male", "female", "male"),
age = c(32, 46, 25, 42, 29))
data1
data2 <- data.frame(id = 6:10,
sex = c("male", "female", "male", "male", "female"),
age = c(52, 36, 28, 34, 26))
data2

rbind(data1, data2)
2. Horizontal merge: cbind ()
To merge two data frames horizontally, you can use cbind( )the function. The two data frames used for merging must have the same number of rows and be in the same order. This kind of merging is often used to add variables to a data frame. For example:
data3 <- data.frame(days = c(28, 57, 15, 7, 19),
outcome = c("discharge", "dead", "discharge", "transfer", "discharge"))
data3
cbind(data1, data3)
3. Merge according to a common variable: merge()
Sometimes we have multiple related data sets that have one or more common variables, and we want to merge them into a large data set based on the common variables. The function merge() can implement this function, for example:
data4 <- data.frame(id = c(2, 1, 3, 5, 4),
outcome = c("discharge", "dead", "discharge", "transfer", "discharge"))
data4
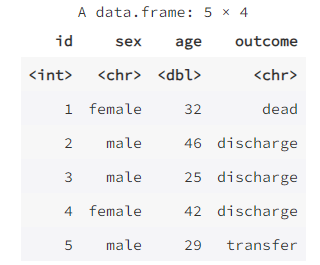
mydata <- merge(data1, data4, by = "id")
mydata
full_join( )
The full_join() function in the dplyr package can also achieve the above function. The above command is equivalent to:
options(warn=-1) # 清爽显示
library(dplyr)
mydata <- full_join(data1, data4, by = "id")
mydata
The dplyr package provides a variety of functions for merging data frames, such as bind_rows(), bind_cols(), left_join(), right_join(), etc. You can check the help documentation of these functions to understand their usage .
4. Conversion of data frame length and width format
Functions in the basic package reshape( )can convert data between long and wide formats.
The following description takes the dataset Indometh in the datasets package as an example. This data set is about the pharmacokinetic data of the drug indometacin. There are a total of 6 test subjects. Each test subject measured the drug concentration in the blood regularly within 8 consecutive hours. A total of 11 times. measured value. The material is in long format, below it is converted to wide format.
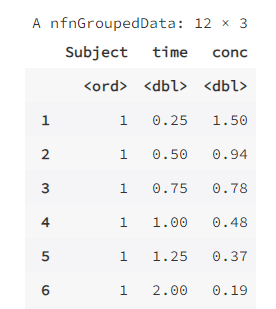
data(Indometh)
head(Indometh,12) # 这里增加一行,预览数据前 12 行,方便对比
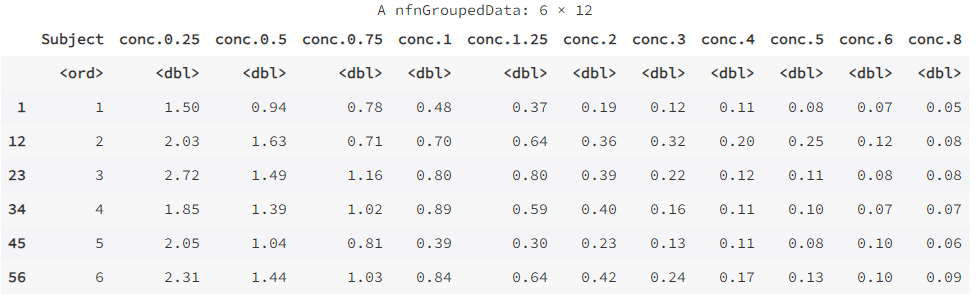
wide <- reshape(Indometh, v.names = "conc", idvar = "Subject", timevar = "time", direction = "wide")
wide
Indometh: This is a data frame or dataset that represents the original data to be subjected to the reshape operation.v.names: This is a string representing the name of the value variable to be reshaped. In this case,"conc"represents the concentration variable in the original data.idvar: This is a string or vector representing a name or list of variables that identifies the variable. In this case,"Subject"represents the subject identification variable in the original data.timevar: This is a string representing the name of the time variable. In this case,"time"represents the time variable in the original data.direction: This is a string indicating the direction of the reshape. In this case,"wide"means that you want to reshape the data from long format to wide format.
We can also re-convert wide format data wide to long format:
long <- reshape(wide, idvar = "Subject", varying = list(2:12),
v.names = "conc", direction = "long")
head(long, 12)

The function reshape() is powerful, but it has many parameters and is a little inconvenient to use.
tidyrThe package implements data long and wide format conversion in a relatively concise and unified format. Among them, the function pivot_wider( )is used to convert long format data to wide format, and the function pivot_longer( )is used to convert wide format data to long format. The above results can also be obtained with the following command:
library(tidyr)
wide <- pivot_wider(as.data.frame(Indometh),
names_from = time,
values_from = conc)
wide
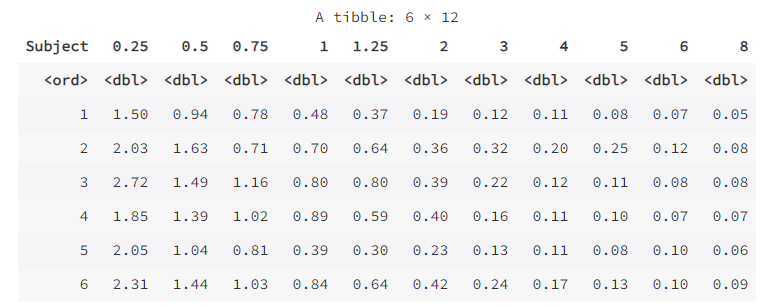
Note that in the function pivot_wider( ) above, we use the function as.data.frame( ) to convert the data Indometh into a data frame because its default type is not a data frame . The data frame wide can also be converted back to long format:
long <- pivot_longer(wide, -Subject,
names_to = "time", values_to = "conc")
long
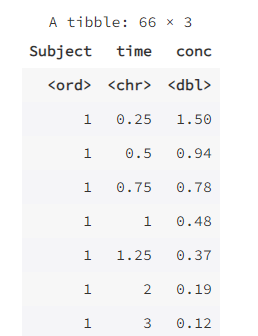
A "tidy" data set (tidy data) should satisfy: each row represents an observation and each column represents a variable. Before analyzing medical data, you should typically convert your data set to long format because most functions in R support data in this format.
gather()and in the tidyr package spread()can also be used for long and wide data type conversion, see Cookbook for R for details .