Article Directory
Environmental description
Anaconda+python3.6+Jupyter Notebook
Anaconda creates a virtual environment and installs the corresponding package
Create a virtual environment
1. Command line creation
Open the command line and
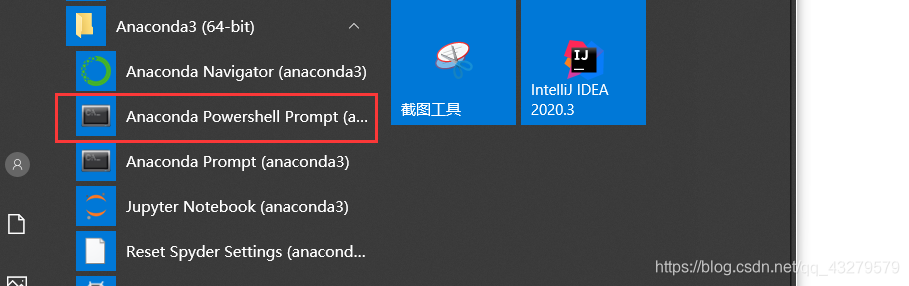
enter the following command
conda create -n sklearn python=3.6
tf1 is the name I chose to create the virtual environment. You can choose the python version according to your needs.
2. Interface creation
Open interface and
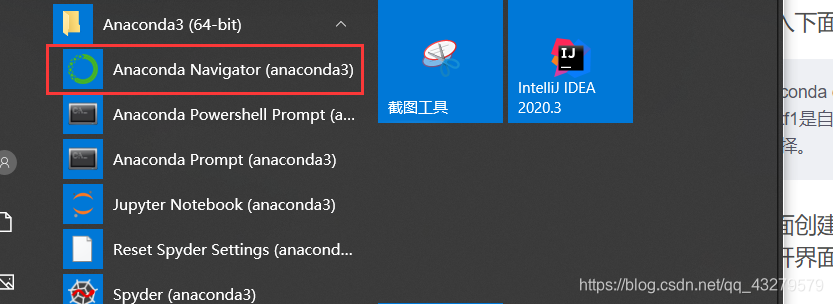
create environment
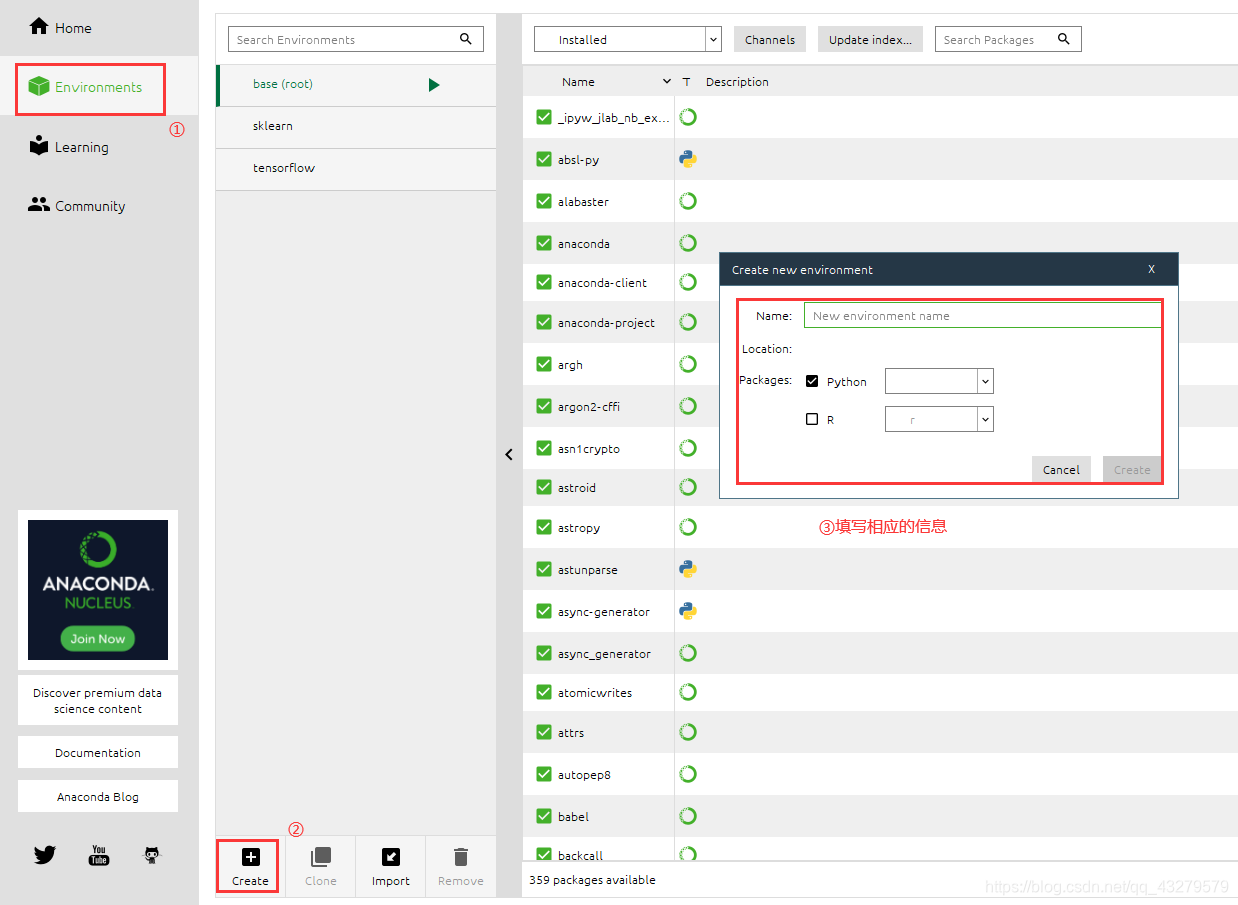
Installation package
pip install package name
Direct installation in this way can cause installation failure or slow installation due to network reasons.
Solution:
pip install -i https://pypi.tuna.tsinghua.edu.cn/simple package name
The package installed here includes numpy , Pandas, sklearn, matplotlib
SVM (Support Vector Machine)
Introduction to SVM
Svm (support Vector Mac), also known as support vector machine, is a two-class model. Support vector machines can be divided into two categories: linear and nonlinear. The main idea is to find a straight line (plane or hyperplane) in the space that is more capable of dividing all data samples, and to make the distance between all data in the data set and this hyperplane the shortest.
The iris data set uses SVM linear classification
LinearSVC (Linear Support Vector Classification) linear support vector machine, the kernel function is linear.
Related parameters:
- C: The penalty coefficient C of the objective function, the default C = 1.0;
- loss: Specify the loss function. squared_hinge (default), squared_hinge
- penalty: penalty method, str type, l1, l2
- dual: Choose an algorithm to solve dual or primitive optimization problems. Dual=false when nsamples>nfeatures
- tol: the precision of the svm end standard, the default is 1e-3
- multi_class: If the y output category contains multiple categories, it is used to determine the multi-category strategy. ovr means one-to-many, and "crammer_singer" optimizes a common goal for all categories. If you choose "crammer_singer", losses, penalties and optimizations will be ignored.
- max_iter: The maximum number of iterations to run. int, default 1000
LinearSVC (C) way to achieve classification
Import the required packages
#导入相应的包
import numpy as np
import matplotlib.pyplot as plt
from sklearn import datasets
from sklearn.preprocessing import StandardScaler
from sklearn.svm import LinearSVC
retrieve data
# 获取所需数据集
iris=datasets.load_iris()
#每行的数据,一共四列,每一列映射为feature_names中对应的值
X=iris.data
#每行数据对应的分类结果值(也就是每行数据的label值),取值为[0,1,2]
Y=iris.target
#通过Y=iris.target.size,可以得到一共150行数据,三个类别个50条数据,并且数据是按照0,1,2的顺序放的
Process the data
#只取y<2的类别,也就是0 1并且只取前两个特征
X=X[:,:2]
#获取0 1类别的数据
Y1=Y[Y<2]
y1=len(Y1)
#获取0类别的数据
Y2=Y[Y<1]
y2=len(Y2)
X=X[:y1,:2]
Plotting of unstandardized raw data points
#绘制出类别0和类别1
plt.scatter(X[0:y2,0],X[0:y2,1],color='red')
plt.scatter(X[y2+1:y1,0],X[y2+1:y1,1],color='blue')
plt.show()

Data normalization processing
#标准化
standardScaler=StandardScaler()
standardScaler.fit(X)
#计算训练数据的均值和方差
X_standard=standardScaler.transform(X)
#用scaler中的均值和方差来转换X,使X标准化
svc=LinearSVC(C=1e9)
svc.fit(X_standard,Y1)
Draw the decision boundary and
the description of the related functions:
- meshgrid() returns a collection of all points in a square space defined by two vectors. x0 is the value of x, x1 is the value of y
- ravel() pulls the vector into a line
- c_[] arrange the vectors together
- contourf() contour
def plot_decision_boundary(model, axis):
x0, x1 = np.meshgrid(
np.linspace(axis[0], axis[1], int((axis[1]-axis[0])*100)).reshape(-1, 1),# 600个,影响列数
np.linspace(axis[2], axis[3], int((axis[3]-axis[2])*100)).reshape(-1, 1),# 600个,影响行数
)
# x0 和 x1 被拉成一列,然后拼接成360000行2列的矩阵,表示所有点
X_new = np.c_[x0.ravel(), x1.ravel()] # 变成 600 * 600行, 2列的矩阵
y_predict = model.predict(X_new) # 二维点集才可以用来预测
zz = y_predict.reshape(x0.shape) # (600, 600)
from matplotlib.colors import ListedColormap
custom_cmap = ListedColormap(['#EF9A9A','#FFF59D','#90CAF9'])
plt.contourf(x0, x1, zz, linewidth=5, cmap=custom_cmap)
#print(X_new)
plot_decision_boundary(svc, axis=[-3, 3, -3, 3])
plt.scatter(X_standard[0:y2,0], X_standard[0:y2,1],color='red')
plt.scatter(X_standard[y2:y1,0], X_standard[y2:y1,1],color='blue')
plt.show()

Instantiate a svc2 (mainly LinearSVC(C)a modification of C)
svc2=LinearSVC(C=0.01)
svc2.fit(X_standard,Y1)
print(svc2.coef_)
print(svc2.intercept_)
plot_decision_boundary(svc2, axis=[-3, 3, -3, 3])
plt.scatter(X_standard[0:y2,0], X_standard[0:y2,1],color='red')
plt.scatter(X_standard[y2:y1,0], X_standard[y2:y1,1],color='blue')
plt.show()

The difference in decision boundary can be clearly seen through the two graphs drawn. The smaller the surface CC, the larger the fault tolerance space.
Add upper and lower boundaries based on the classified content
def plot_svc_decision_boundary(model, axis):
x0, x1 = np.meshgrid(
np.linspace(axis[0], axis[1], int((axis[1]-axis[0])*100)).reshape(-1, 1),# 600个,影响列数
np.linspace(axis[2], axis[3], int((axis[3]-axis[2])*100)).reshape(-1, 1),# 600个,影响行数
)
# x0 和 x1 被拉成一列,然后拼接成360000行2列的矩阵,表示所有点
X_new = np.c_[x0.ravel(), x1.ravel()] # 变成 600 * 600行, 2列的矩阵
y_predict = model.predict(X_new) # 二维点集才可以用来预测
zz = y_predict.reshape(x0.shape) # (600, 600)
from matplotlib.colors import ListedColormap
custom_cmap = ListedColormap(['#EF9A9A','#FFF59D','#90CAF9'])
plt.contourf(x0, x1, zz, linewidth=5, cmap=custom_cmap)
w = model.coef_[0]
b = model.intercept_[0]
index_x = np.linspace(axis[0], axis[1], 100)
# f(x,y) = w[0]x1 + w[1]x2 + b
# 1 = w[0]x1 + w[1]x2 + b 上边界
# -1 = w[0]x1 + w[1]x2 + b 下边界
y_up = (1-w[0]*index_x - b) / w[1]
y_down = (-1-w[0]*index_x - b) / w[1]
x_index_up = index_x[(y_up<=axis[3]) & (y_up>=axis[2])]
x_index_down = index_x[(y_down<=axis[3]) & (y_down>=axis[2])]
y_up = y_up[(y_up<=axis[3]) & (y_up>=axis[2])]
y_down = y_down[(y_down<=axis[3]) & (y_down>=axis[2])]
plt.plot(x_index_up, y_up, color="black")
plt.plot(x_index_down, y_down, color="black")
plot_svc_decision_boundary(svc, axis=[-3, 3, -3, 3])
plt.scatter(X_standard[0:y2,0], X_standard[0:y2,1],color='red')
plt.scatter(X_standard[y2:y1,0], X_standard[y2:y1,1],color='blue')
plt.show()

Modify the value of C
plot_svc_decision_boundary(svc2, axis=[-3, 3, -3, 3])
plt.scatter(X_standard[0:y2,0], X_standard[0:y2,1],color='red')
plt.scatter(X_standard[y2:y1,0], X_standard[y2:y1,1],color='blue')
plt.show()

Conclusion: The
larger the constant C, the smaller the fault-tolerant space and the closer the upper and lower boundaries; the smaller the constant C, the larger the fault-tolerant space and the farther the upper and lower boundaries.