Neo4j is an open source, high-performance graph database. It is designed to store, retrieve and process large-scale data with complex relationships. Different from traditional relational databases, Neo4j uses a graph structure to represent data, where nodes represent entities and edges represent relationships between entities. This makes Neo4j very powerful and efficient when dealing with relationship-intensive data.
Here are some of the key features of Neo4j:
-
Graph database: Neo4j is a native, completely graph-based database that provides a graph model to store and manage data. This enables it to easily handle complex data relationships such as social networks, recommendation systems, cybersecurity, etc.
-
Flexible data model: Neo4j’s data model is very flexible and can easily represent various types of entities and relationships. You can define your own node labels and edge types and add properties to them to better represent your data structure.
-
High performance: Neo4j achieves excellent performance by using efficient graph traversal algorithms and indexing mechanisms. It can quickly perform complex graph queries, supporting deep traversal and fast relationship navigation.
-
Native query language: Neo4j uses a native query language called Cypher to manipulate and query graph data. Cypher has an intuitive syntax that can easily express various graph query patterns, including node and edge matching, path traversal, and aggregation operations.
-
ACID transaction support: Neo4j supports atomicity, consistency, isolation, and durability (ACID) transaction features. This ensures data integrity and consistency while providing a reliable persistence mechanism.
-
Ecosystem and tool support: Neo4j has a rich ecosystem and tool support, including visualization tools, ETL tools, drivers and integration libraries, etc. These tools make it easier for developers and data analysts to use and operate Neo4j databases.
Neo4j official website is here , as shown below:
You can download, install and use it according to your own needs.
The screenshot of the startup after the installation is completed is as follows:
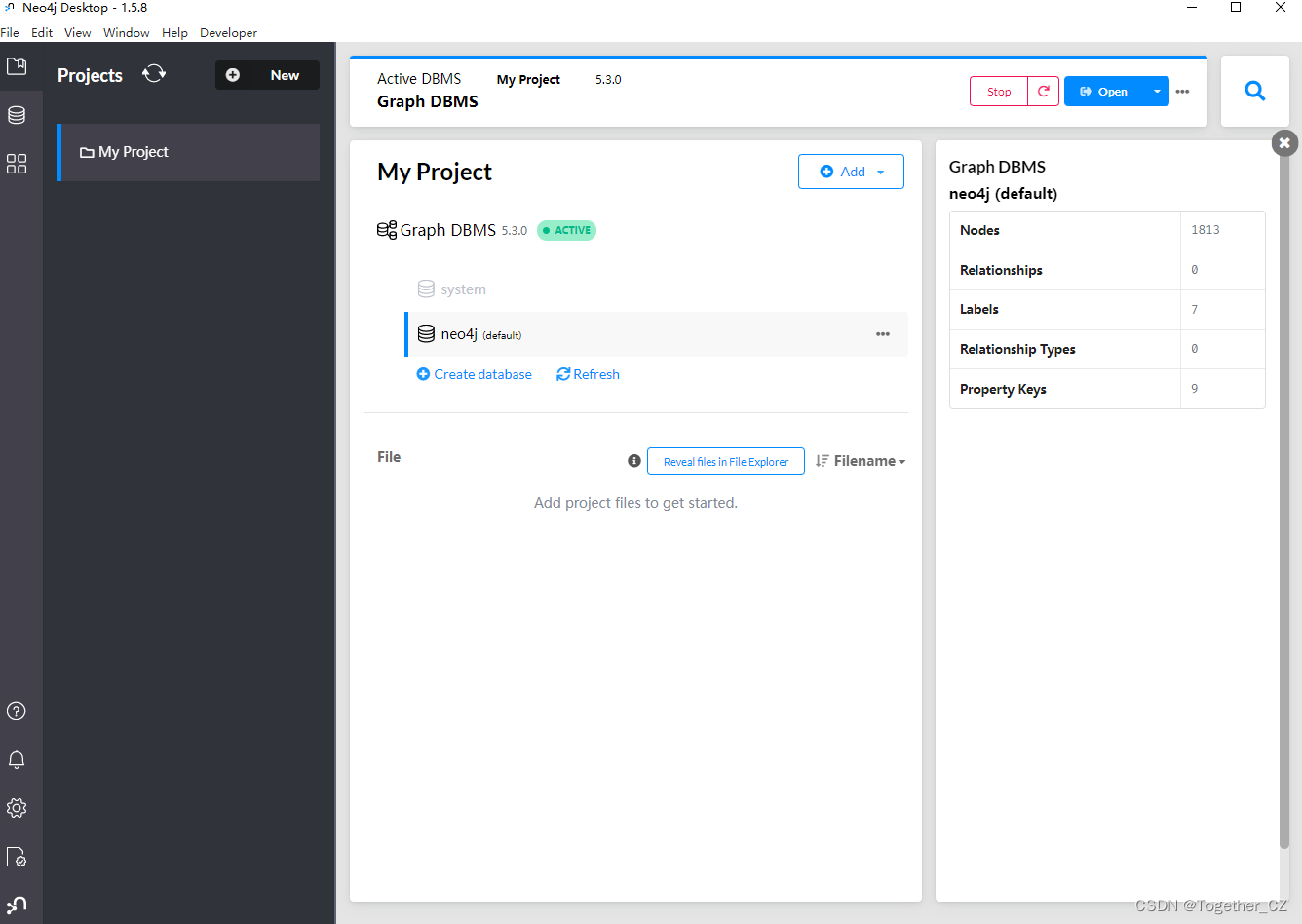
What I installed here is version 1.5.8.
I had come into contact with the Neo4j database when I was in school. I just installed it and practiced it briefly. Later, because the subject direction changed, I didn’t come into contact with it again. I happened to see this graph database again recently when I had nothing to do on the weekend. Content, just think about it
Let’s get started with a practical development project. Here we mainly use the pig-related breeding disease data set collected in the previous project as a benchmark to build a question and answer system based on the knowledge graph.
Knowledge graph is a graphical knowledge base used to organize, represent and store structured knowledge. It represents real-world entities, concepts and the relationships between them as graphical structures, which can help us better understand and analyze knowledge.
Knowledge graphs usually consist of three main components:
-
Entities: Entities represent specific things in the real world, which can be people, places, items, events, etc. Each entity has a unique identifier and associated attribute information.
-
Attributes: Attributes describe the characteristics and properties of an entity. For example, a person entity can have attributes such as name, age, gender, etc.
-
Relationships: Relationships represent connections and associations between entities. They describe semantic relationships between entities, such as "works in", "is located in", "is a subset/superset", etc.
The advantage of the knowledge graph is that it can capture and represent complex relationships and provide a structured way to store and query knowledge. It can be used to build intelligent recommendation systems, question and answer systems, semantic search engines and other applications. By using knowledge graphs, we can perform more advanced data analysis, semantic reasoning, and information extraction.
For example, taking an encyclopedia as an example, a knowledge graph can organize entities such as different topics, characters, events, etc. into a graphic structure and link them using relationships. In this way, we can easily browse and discover related knowledge, such as finding family members of a certain character, understanding the historical background of an event, and so on.
First, let’s take a brief look at the data set, as shown below:
"猪附红细胞体病": {
"name": "猪附红细胞体病",
"desc": "由附红细胞体寄生于猪的红细胞表面或游离于血浆、组织液及脑脊液中引起的一种人畜共患病,猪发病时,皮肤发红,故又称“猪红皮病”。",
"cause": "附红细胞体病是由多种原因引发的疾病,只有在应激和肌体抗病力降低的情况下才会诱发此病。如饲养管理不良、天气突变、突然换料、更换圈舍、密度过大等应激因素或患猪瘟、猪蓝耳病、传染性胸膜肺炎、猪链球菌病、副猪嗜血杆菌病等慢性病时,最易并发和继发附红细胞体病。",
"prevent": "预防本病的发生应加强猪场的卫生防疫,消除各种应激因素。在温热季节应定期喷洒杀虫剂,以杀灭蚊、蝇、蜱、牛虻、体虱、跳蚤等吸血昆虫,消除传染媒介。发病猪只要进行及时有效的治疗。对无治疗价值的病猪应及时进行淘汰,以清除传染源。阳性猪群,饲料中可添加强力霉素等,以消除隐性感染。购入猪只要进行血液检查,防止引入病猪或隐性感染猪。本病流行季节给予预防用药,可在饲料中添加上强力霉素或金霉素添加剂,或每公斤饲料添加90毫克阿散酸,连续使用30天,或每月使用7-10天。 防重于治是养猪的最后方法。",
"cure_lasttime": "一般3-14天。",
"cure_way": "⑴血虫净(或三氮眯、贝尼尔) 每公斤体重用5mg-10mg,用生理盐水稀释成5%溶液,分点肌肉注射,1天1次,连用3天。⑵咪唑苯脲每公斤体重用1mg-3mg,1天1次,连用2天-3天。⑶四环素、土霉素(每公斤体重10mg)和金霉素(每公斤体重15mg)口服或肌注或静注,连用7天-14天。⑷新胂凡纳明按每公斤体重10mg-15mg静脉注射,一般3天后症状可消失。",
"easy_get": "不同年龄的猪均有易感性,通常发生在哺乳猪、怀孕的母猪以及受到高度应激的肥育猪身上。",
"symptom": [
"体温升高为40.5℃~42℃",
"皮肤发红,指压退色",
"精神不振",
"食欲减退",
"怕冷聚堆",
"咳嗽",
"流鼻涕",
"呼吸困难",
"尿液淡黄",
"发病中期,病猪行走时后躯摇晃,喜卧厌立,便秘或拉稀,精神沉郁,呼吸困难",
"血液稀薄,色淡,往往随注射针孔流血不止",
"皮毛枯燥",
"背腹部毛色铁锈色",
"皮肤苍白",
"耳内侧、背侧、颈背部、腹侧部皮肤出现暗红色出血点,可视黏膜轻度肿胀,初期潮红,后期苍白",
"轻度黄疸",
"尿液淡黄、淡红或呈红褐色,卧地不起",
"后期,病猪耳朵变蓝色、坏死,排血便和血红蛋白尿,最后四肢呈游泳状划动,呼吸困难,衰竭死亡"
],
"recommand_drug": [
"抗生素",
"磺胺类",
"砷制剂",
"血虫净",
"三氮眯",
"贝尼尔",
"咪唑苯脲",
"四环素",
"土霉素",
"金霉素",
"新胂凡纳明"
],
"checks": [
"猪附红细胞体病的发热、贫血、黄疸等症状具有一定的诊断意义,其他临床症状,如食欲减退、呼吸急迫、心悸亢进等非特征性症状在本病的诊断上意义不大。",
"猪患附红细胞体病后全身各部均无特征性的病理变化,一般无需作病理学检查。因此,对本病确诊,必须先进行实验室检查。",
"实验室诊断的方法很多,如可用间接红细胞凝集试验、补体结合试验、相差显微镜观察和染色血液涂片观察等。",
"间接红细胞凝集试验和补体结合试验反应敏感、检出率高,但试验条件要求高,需要时间较长。",
"相差显微镜观察和染色血液涂片观察以及鲜血直接压片,所需设备和药品较少,且操作简单、快捷,检出率也较高,一般在半个小时内即可作出确切诊断。"
],
"departments": [
"寄生虫病"
],
"methods": [
"温热季节定期喷洒杀虫剂",
"及时治疗发病猪只,无治疗价值的猪只应及时淘汰",
"阳性猪群,饲料中可添加强力霉素等,以消除隐性感染。",
"购入猪只要进行血液检查,防止引入病猪或隐性感染猪。"
],
"acompany_with": [
"链球菌病",
"猪水肿病",
"仔猪副伤寒",
"猪肺疫",
"猪丹毒",
"猪瘟",
"弓形虫病"
]
}The above is the relevant content details of a single disease. You can see that it contains a lot of content.
The following is an introduction to some common pig diseases, as follows:
{
"猪附红细胞体病": {
"疾病所属类别": "寄生虫病",
"疾病诊断要点": "相差显微镜观察和染色血液涂片观察以及鲜血直接压片,所需设备和药品较少,且操作简单、快捷,检出率也较高,一般在半个小时内即可作出确切诊断。",
"疾病推荐药物": "新胂凡纳明",
"疾病预防措施": "购入猪只要进行血液检查,防止引入病猪或隐性感染猪。"
},
"副猪嗜血杆菌病": {
"疾病所属类别": "细菌性传染病",
"疾病诊断要点": "该病易与传染性胸膜肺炎相混淆,但该病引起的病变多数为脑膜炎,关节炎和四肢跛行等,而传染性胸膜性肺炎较少见。",
"疾病推荐药物": "阿莫西林",
"疾病预防措施": "疫苗免疫"
},
"猪支原体肺炎": {
"疾病所属类别": "传染病",
"疾病诊断要点": "其他内脏一般无明显变化。",
"疾病推荐药物": "中药方剂 (苏子、杏仁、款冬花、桔梗、甘草、陈皮、鱼腥草等)",
"疾病预防措施": "加强消毒,保持栏舍清洁、干燥通风。"
},
"猪圆环病毒病": {
"疾病所属类别": "病毒性传染病",
"疾病诊断要点": "血清学检查:是生前诊断的一种有效手段。诊断本病的方法有:间接免疫荧光法(IIF),免疫过氧化物单层培养法,ELISA方法,聚合酶链式反应(PCR)方法,核酸探针杂交及原位杂交试验(ISH)等方法。",
"疾病推荐药物": "选用新型的抗病毒剂如干扰素、白细胞介导素、免疫球蛋白、转移因子等进行治疗,同时配合中草药抗病毒制剂,会取得明显治疗效果。",
"疾病预防措施": "加强饲养管理"
},
"猪链球菌病": {
"疾病所属类别": "人畜共患病",
"疾病诊断要点": "药敏试验",
"疾病推荐药物": "抗生素",
"疾病预防措施": "加强饲养管理"
},
"猪伪狂犬病": {
"疾病所属类别": "急性传染病",
"疾病诊断要点": "血清学诊断可直接用免疫荧光法、间接血凝抑制试验、琼脂扩散试验、补体结合试验、酶联免疫吸附试验、乳胶凝集试验。",
"疾病推荐药物": "猪血清抗体",
"疾病预防措施": "同时,还要严格控制犬、猫、鸟类和其他禽类进入猪场,严格控制人员来往,并做好消毒工作及血清学监测等,这样对本病的防制也可起到积极的推动作用。"
}Next, you need to develop a program to complete the data loading, parsing and storage operations. Here, the pigMedicalGraph class is defined as follows:
#!usr/bin/env python
# encoding:utf-8
from __future__ import division
"""
功能: 猪类疾病知识图谱构建
"""
import os
import json
from py2neo import Graph,Node
class pigMedicalGraph:
def __init__(self):
def read_nodes(self):
def create_node(self, label, nodes):
def create_diseases_nodes(self, disease_infos):
def create_graphnodes(self):
def create_graphrels(self):
def create_relationship(self, start_node, end_node, edges, rel_type, rel_name):
The first is to load and parse local data, as shown below:
count = 0
with open(self.data_path, encoding="utf-8") as f:
data_dict = json.load(f)
for one_key in data_dict:
data_json = data_dict[one_key]
disease_dict = {}
count += 1
print(count)
disease = data_json["name"]
print(disease)
disease_dict["name"] = disease
if type(disease) == list:
diseases += disease
disease = disease[0]
else:
diseases.append(disease)
disease_dict["desc"] = ""
disease_dict["cause"] = ""
disease_dict["prevent"] = ""
disease_dict["cure_lasttime"] = ""
disease_dict["cure_way"] = ""
disease_dict["symptom"] = ""
disease_dict["recommand_drug"] = ""
print("59")
if "symptom" in data_json:
symptoms += data_json["symptom"]
for symptom in data_json["symptom"]:
has_symptom.append([disease, symptom])
if "checks" in data_json:
checks += data_json["checks"]
for check in data_json["checks"]:
need_check.append([disease, check])
if "departments" in data_json:
departments += data_json["departments"]
for department in data_json["departments"]:
belongs_to.append([disease, department])
if "methods" in data_json:
methods += data_json["methods"]
for method in data_json["methods"]:
recommand_method.append([disease, method])
if "desc" in data_json:
disease_dict["desc"] = data_json["desc"]
if "prevent" in data_json:
disease_dict["prevent"] = data_json["prevent"]
if "cause" in data_json:
disease_dict["cause"] = data_json["cause"]
if "easy_get" in data_json:
disease_dict["easy_get"] = data_json["easy_get"]
if "cure_way" in data_json:
disease_dict["cure_way"] = data_json["cure_way"]
if "cure_lasttime" in data_json:
disease_dict["cure_lasttime"] = data_json["cure_lasttime"]
if "recommand_drug" in data_json:
recommand_drug1 = data_json["recommand_drug"]
drugs += recommand_drug1
for drug in recommand_drug1:
recommand_drug.append([disease, drug])
disease_infos.append(disease_dict)
The next step is to create the node node in the knowledge graph. The implementation is as follows:
def create_node(self, label, nodes):
"""
创建节点
"""
count = 0
for node_name in nodes:
node = Node(label, name=node_name)
self.g.create(node)
count += 1
print(count, len(nodes))
return
Then we will implement the creation of disease nodes in the knowledge graph, as shown below:
def create_diseases_nodes(self, disease_infos):
"""
创建知识图谱中疾病的节点
"""
count = 0
for disease_dict in disease_infos:
print(disease_dict)
node = Node(
"Disease",
name=disease_dict["name"],
desc=disease_dict["desc"],
cause=disease_dict["cause"],
prevent=disease_dict["prevent"],
cure_lasttime=disease_dict["cure_lasttime"],
cure_way=disease_dict["cure_way"],
easy_get=disease_dict["easy_get"],
recommand_drug=disease_dict["recommand_drug"],
symptom=disease_dict["symptom"],
)
print("132")
self.g.create(node)
count += 1
print(count)
return
The next step is to create the knowledge graph entity node type schema. The core implementation is as follows:
self.create_diseases_nodes(disease_infos)
self.create_node("checks", checks)
self.create_node("departments", departments)
self.create_node("diseases", diseases)
self.create_node("drugs", drugs)
self.create_node("methods", methods)
self.create_node("symptoms", symptoms)Then we create the entity association edge, as shown below:
def create_relationship(self, start_node, end_node, edges, rel_type, rel_name):
"""
创建实体关联边
"""
count = 0
# 去重处理
set_edges = []
for edge in edges:
set_edges.append("".join(edge))
all = len(set(set_edges))
for edge in set(set_edges):
edge = edge.split("")
p = edge[0]
q = edge[1]
query = (
"match(p:%s),(q:%s) where p.name='%s' and q.name='%s' create (p)-[rel:%s{name:'%s'}]->(q)"
% (start_node, end_node, p, q, rel_type, rel_name)
)
try:
self.g.run(query)
count += 1
print(rel_type, count, all)
except Exception as e:
print(e)
return
Finally we create the entity relationship edge, as shown below:
self.create_relationship("diseases", "departments", belongs_to, "belongs_to", "属于")
self.create_relationship("diseases", "checks", need_check, "need_check", "疾病诊断结果")
self.create_relationship(
"diseases", "drugs", recommand_drug, "recommand_drug", "疾病推荐药品"
)
self.create_relationship(
"diseases", "methods", recommand_method, "recommand_method", "疾病预防措施"
)
self.create_relationship("diseases", "symptoms", has_symptom, "has_symptom", "疾病症状")
self.create_relationship(
"diseases", "diseases", acompany_with_, "acompany_with_", "疾病并发疾病"
)
At this point, the data analysis and loading operation is basically completed.
You can open and view it directly on the neo4j database desktop, as shown below:
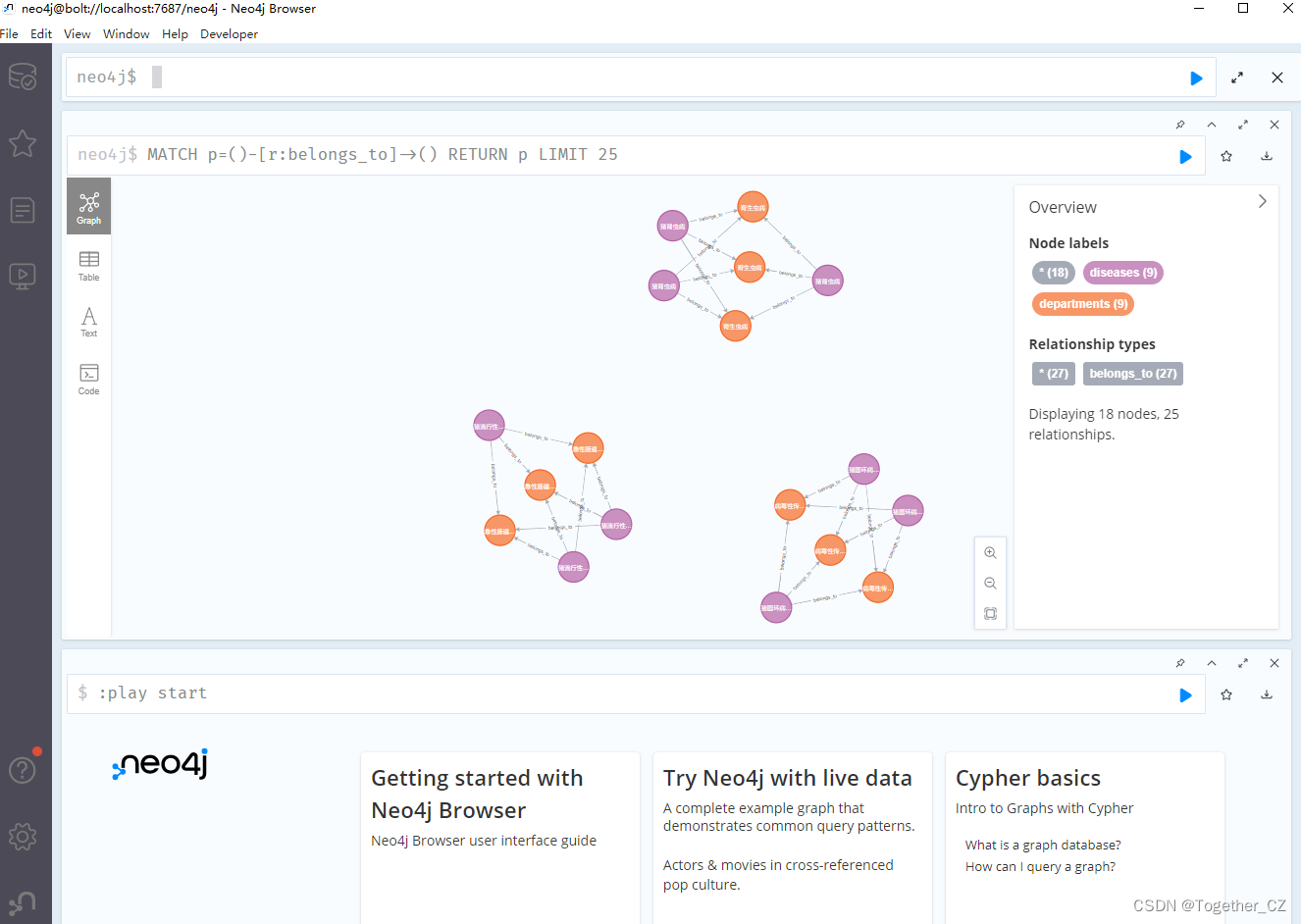
You can also directly copy http://localhost:7474/browser/
Just open it on the browser, as shown below:
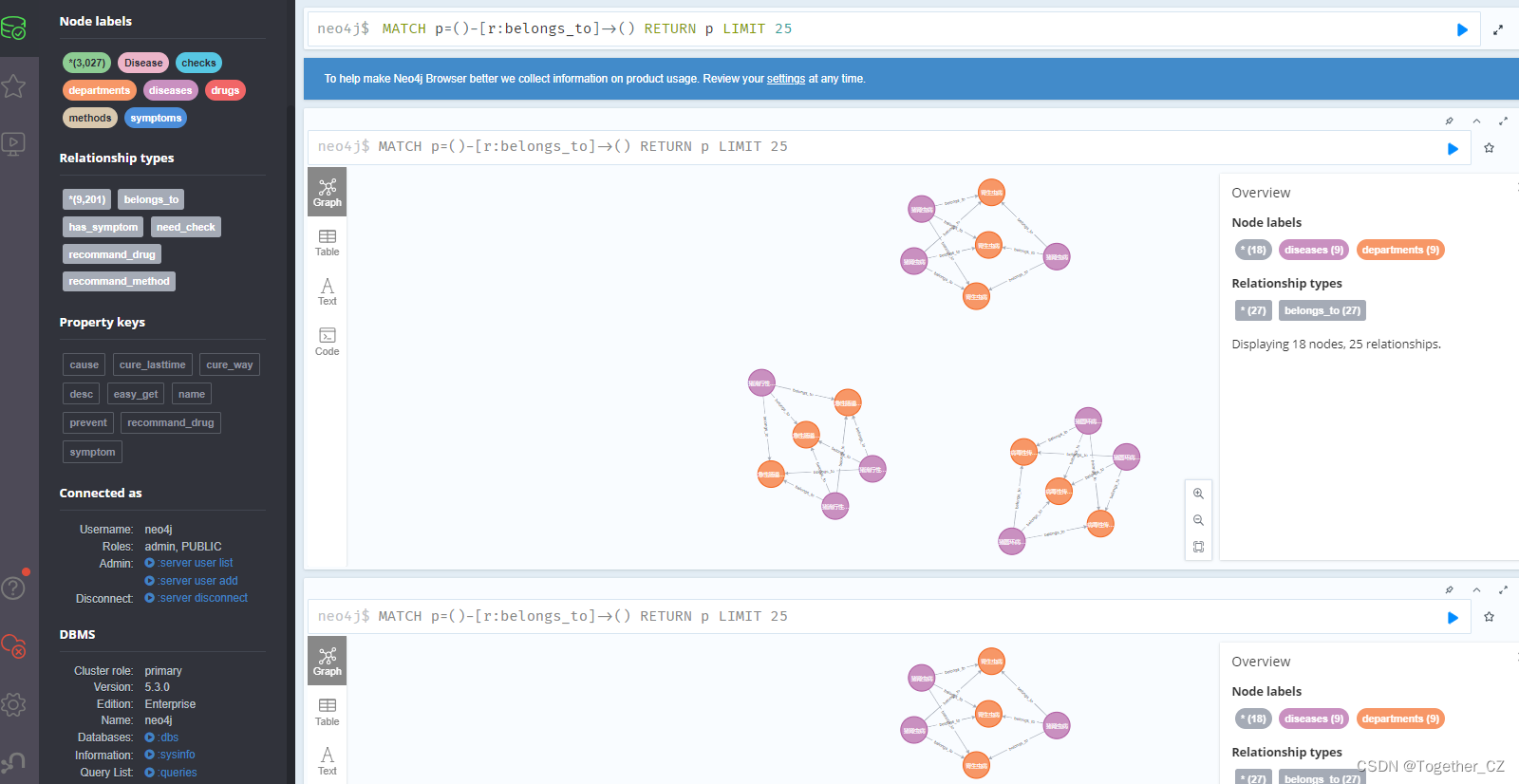
Obviously I feel that the operations and visualization on the browser side are smoother and smoother.
Here you can directly use the ability of the knowledge graph to answer knowledge queries. The simple implementation is as follows:
class ChatRobot:
def __init__(self):
self.classifier = QuestionClassifier()
self.parser = QuestionPaser()
self.searcher = AnswerSearcher()
def chat_main(self, sent):
answer = "尊敬的用户您好,我是AI医药智能助理,希望可以帮到您!"
res_classify = self.classifier.classify(sent)
if not res_classify:
return answer
res_sql = self.parser.parser_main(res_classify)
print("res_sql: ", res_sql)
final_answers = self.searcher.search_main(res_sql)
if not final_answers:
return answer
else:
return "\n".join(final_answers)
if __name__ == "__main__":
handler = ChatRobot()
while True:
question = input("user:")
answer = handler.chat_main(question)
print("AI:", answer)
Example output is as follows:

Next, let’s take a brief look at the visual presentation of the neo4j database:
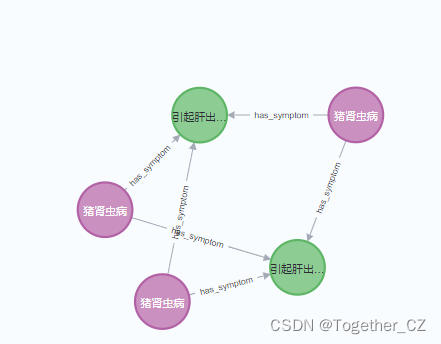

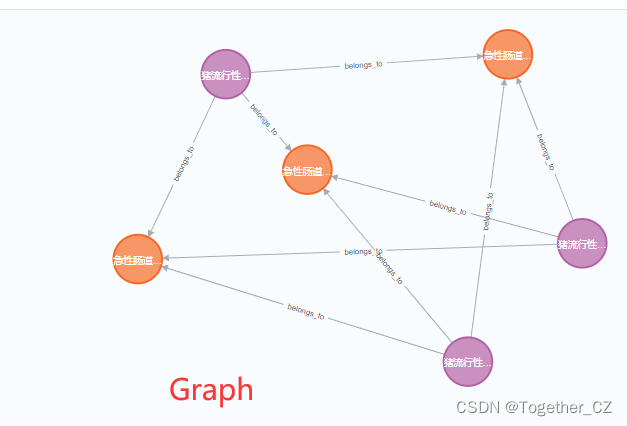
Each block includes four representations or storage forms: Graph, Table, Text and Code. Next, we will show it with an actual example, as shown below:
【Graph】
【Table】
{
"start": {
"identity": 284,
"labels": [
"diseases"
],
"properties": {
"name": "猪圆环病毒病"
},
"elementId": "284"
},
"end": {
"identity": 230,
"labels": [
"departments"
],
"properties": {
"name": "病毒性传染病"
},
"elementId": "230"
},
"segments": [
{
"start": {
"identity": 284,
"labels": [
"diseases"
],
"properties": {
"name": "猪圆环病毒病"
},
"elementId": "284"
},
"relationship": {
"identity": 0,
"start": 284,
"end": 230,
"type": "belongs_to",
"properties": {
"name": "属于"
},
"elementId": "0",
"startNodeElementId": "284",
"endNodeElementId": "230"
},
"end": {
"identity": 230,
"labels": [
"departments"
],
"properties": {
"name": "病毒性传染病"
},
"elementId": "230"
}
}
],
"length": 1.0
}【Text】
╒════════════════════════════════════════════════════╕
│"p" │
╞════════════════════════════════════════════════════╡
│[{"name":"猪圆环病毒病"},{"name":"属于"},{"name":"病毒性传染病"}] │
├────────────────────────────────────────────────────┤
│[{"name":"猪圆环病毒病"},{"name":"属于"},{"name":"病毒性传染病"}] │
├────────────────────────────────────────────────────┤
│[{"name":"猪圆环病毒病"},{"name":"属于"},{"name":"病毒性传染病"}] │
├────────────────────────────────────────────────────┤
│[{"name":"猪圆环病毒病"},{"name":"属于"},{"name":"病毒性传染病"}] │
├────────────────────────────────────────────────────┤
│[{"name":"猪圆环病毒病"},{"name":"属于"},{"name":"病毒性传染病"}] │
├────────────────────────────────────────────────────┤
│[{"name":"猪圆环病毒病"},{"name":"属于"},{"name":"病毒性传染病"}] │
├────────────────────────────────────────────────────┤
│[{"name":"猪圆环病毒病"},{"name":"属于"},{"name":"病毒性传染病"}] │
├────────────────────────────────────────────────────┤
│[{"name":"猪圆环病毒病"},{"name":"属于"},{"name":"病毒性传染病"}] │
├────────────────────────────────────────────────────┤
│[{"name":"猪圆环病毒病"},{"name":"属于"},{"name":"病毒性传染病"}] │
├────────────────────────────────────────────────────┤
│[{"name":"猪肾虫病"},{"name":"属于"},{"name":"寄生虫病"}] │
├────────────────────────────────────────────────────┤
│[{"name":"猪肾虫病"},{"name":"属于"},{"name":"寄生虫病"}] │
├────────────────────────────────────────────────────┤
│[{"name":"猪肾虫病"},{"name":"属于"},{"name":"寄生虫病"}] │
├────────────────────────────────────────────────────┤
│[{"name":"猪肾虫病"},{"name":"属于"},{"name":"寄生虫病"}] │
├────────────────────────────────────────────────────┤
│[{"name":"猪肾虫病"},{"name":"属于"},{"name":"寄生虫病"}] │
├────────────────────────────────────────────────────┤
│[{"name":"猪肾虫病"},{"name":"属于"},{"name":"寄生虫病"}] │
├────────────────────────────────────────────────────┤
│[{"name":"猪肾虫病"},{"name":"属于"},{"name":"寄生虫病"}] │
├────────────────────────────────────────────────────┤
│[{"name":"猪肾虫病"},{"name":"属于"},{"name":"寄生虫病"}] │
├────────────────────────────────────────────────────┤
│[{"name":"猪肾虫病"},{"name":"属于"},{"name":"寄生虫病"}] │
├────────────────────────────────────────────────────┤
│[{"name":"猪流行性腹泻"},{"name":"属于"},{"name":"急性肠道传染病"}]│
├────────────────────────────────────────────────────┤
│[{"name":"猪流行性腹泻"},{"name":"属于"},{"name":"急性肠道传染病"}]│
├────────────────────────────────────────────────────┤
│[{"name":"猪流行性腹泻"},{"name":"属于"},{"name":"急性肠道传染病"}]│
├────────────────────────────────────────────────────┤
│[{"name":"猪流行性腹泻"},{"name":"属于"},{"name":"急性肠道传染病"}]│
├────────────────────────────────────────────────────┤
│[{"name":"猪流行性腹泻"},{"name":"属于"},{"name":"急性肠道传染病"}]│
├────────────────────────────────────────────────────┤
│[{"name":"猪流行性腹泻"},{"name":"属于"},{"name":"急性肠道传染病"}]│
├────────────────────────────────────────────────────┤
│[{"name":"猪流行性腹泻"},{"name":"属于"},{"name":"急性肠道传染病"}]│
└────────────────────────────────────────────────────┘【Code】
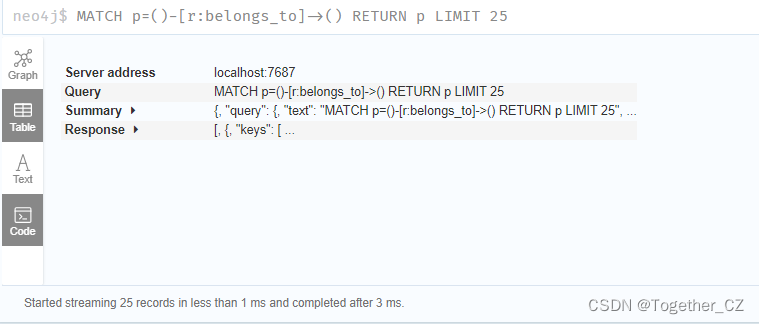
This code can be copied directly and executed in the Neo4j desktop to get the same results as the browser page.
If you are interested, you can also try to develop and build your own application system!