LabVIEW Using Support Vector Machines to Classify Brain MRI Images
Medical imaging is the technology and process used to create images of human anatomy for clinical research, diagnosis and treatment. It is now one of the fastest growing areas of medical technology. The modalities commonly used to obtain medical images are X-rays, computed tomography (CT), magnetic resonance imaging (MRI), and ultrasound imaging. In medical imaging, an MRI is one of the scanning devices that uses a magnetic field to capture images onto film. Due to its excellent soft tissue contrast and detailed resolution, MRI is used for anatomical assessment of human brain structures.
Brain MRI images may contain normal or defective abnormal slices. Normal and abnormal brain images are determined by their symmetry on axial and coronal images. Asymmetry beyond a certain degree is a clear sign of a diseased brain, which has been exploited in our work for preliminary classification at a coarse level. Therefore, further examination of the images involving MRI brain classification is required.
The purpose of classification is to group items with similar feature values into categories. Many studies have been conducted on brain MRI classification. These include techniques using neural networks, fuzzy c-means, k-nearest neighbors, and support vector machines (SVM).
SVM has been widely used in pattern recognition applications due to its computational efficiency and good generalization performance. There is also a need to improve the process before a patient's condition can be determined based on Alzheimer's disease. This is to determine the likelihood that an image falls into the normal or abnormal category based on symmetry, as is the case with Alzheimer's disease or brain tumors. In this work, two classes of image classification, normal or abnormal, are automatically obtained using SVM.
The key concept of SVM is the use of hyperplanes to define decision boundaries between data points that separate different classes. SVMs can handle both simple linear classification tasks and more complex (i.e. non-linear) classification problems. Both separable and non-separable problems are handled by SVMs in both linear and nonlinear cases. The idea behind SVM is to map the raw data points from the input space to a high-dimensional or even infinite-dimensional feature space, thus making classification problems in the feature space much simpler. Mapping is done by appropriate selection of kernel functions.
Images are enhanced using image enhancement techniques such as image denoising and noise filtering. First apply the wavelet transform to the image. The obtained wavelet coefficients are then manipulated to remove noise points. The basis of the compressed or localized wavelet transform is called the mother wavelet of the wavelet transform.
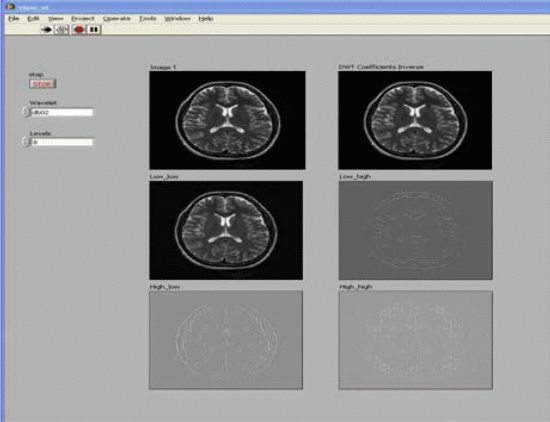
The starting point of the project is to use the wavelet approximation coefficients of brain MRI as input to the SVM. Through machine learning methods, it is hoped to achieve greater precision and accuracy in interpreting normal and abnormal brain images.