Matrix multiplication calculation GPU implementation usually calculates a larger [m_tile, k] *[k, n_tile] matrix multiplication for thread blocks, and finally assigns to each thread and also calculates a smaller one for each thread [m_tile, k] *[k, n_tile].
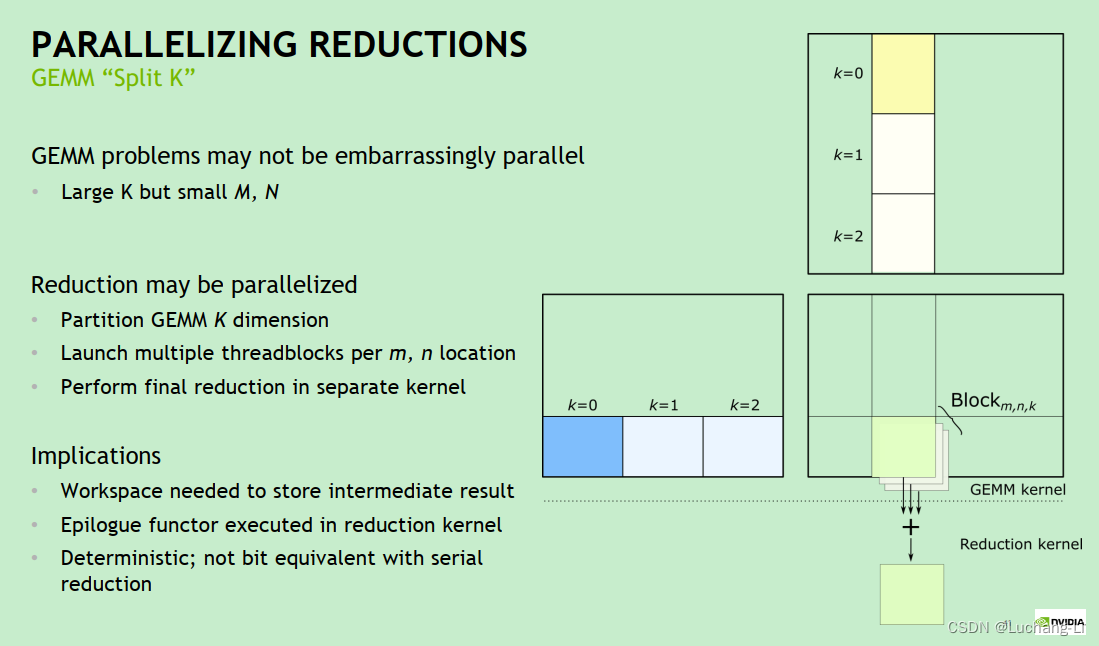
One problem with this is that when m and n are small and k is large, only a few threads and thread blocks can be allocated in the case of matrix multiplication as shown in the figure below, and the number of loops inside each thread is large. The GPU cannot be fully utilized, resulting in poor performance of the matrix multiplication implementation. This situation may widely appear in matrix multiplication obtained by convolution through im2col/im2row method conversion: Convolution optimization techniques in OpenPPL-Know almost

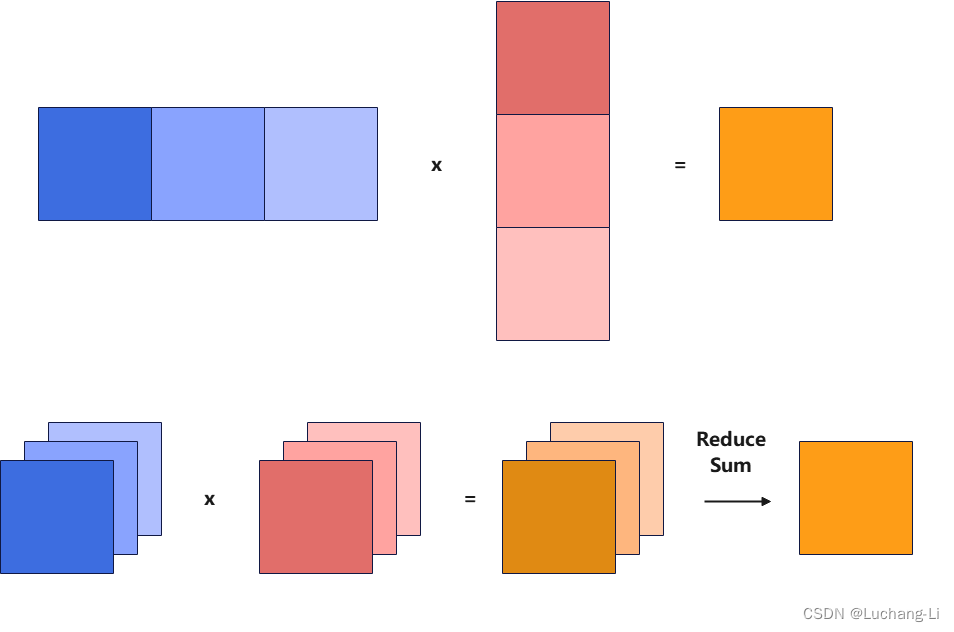
The principle of splitk is to split the k-direction of matrix multiplication into multiple k_n smaller k_sizes, thus obtaining k_n [m, k_tile] x [k_tile, n] matrix multiplications, and the k loop size of each matrix multiplication is shortened. Thus, the calculation time per thread is shortened, and a higher number of threads can be created to perform calculations.
The basic principle is shown in the figure below, that is, multiple matrix multiplications with smaller k are calculated in parallel, and an additional ReduceSum operator is added for cumulative calculation.
Is there an easy way to achieve the above optimization?
The answer is that it can be optimized through a very simple and general graph without the need to add or modify the existing operator implementation of the inference engine, but the performance may be slightly worse than the specially implemented splitk matrix multiplication.
Assume that the shape of the matrix multiplied by input a is [Ba, M, K]. Ba is the batch of input a, which can be any number of dimensions. Now first perform a reshape to get [Ba, M, Kn, K0], and then perform a transpose to get [Ba, Kn, M, K0], then you can get the matrix multiplied by the new input a after splitk.
Similarly, the shape of the matrix multiplied by input b is [Bb, K, N]. Bb is the batch of input b, which can be any number of dimensions. Now perform reshape to get [Bb, Kn, K0, N], which is the new input b multiplied by the matrix after splitk.
Then the batch matrix multiplication of [Ba, Kn, M, K0] and [Bb, Kn, K0, N] achieves the effect of split k. Finally, insert a ReduceSum(axis=-3) after the matrix multiplication operator to complete.
This graph optimization inserts two reshapes, one transpose, and one reduce. Reduce is inevitable, and the reshape operator is actually just memory reinterpretation, and does not need to be time-consuming for real calculations. Therefore, compared with the special splitk matrix multiplication, it takes one more transpose time-consuming. Of course, the time-consuming of this operator is much lower than the time-consuming of matrix multiplication.
One thing to note is the bias of matrix multiplication. When converting splitk to batch matrix multiplication above, if there is a bias, it will cause a bias to be added to each batch of k splits, but only one bias should be added. . One solution is to split bias into an add operator and add it after reduceSum. This add can be integrated with other elemwise behind matmul to reduce performance loss.
The performance benefit of this method on the NV GPU may not be as high as that of the end-side GPU, because the end-side GPU is difficult to use shared mem to accelerate, but the method in this article may be a good method.
numpy reference code
import numpy as np
shape_a = [1, 49, 2016]
shape_b = [2016, 448]
np.random.seed(1)
data_a = np.random.uniform(-1, 1, size=shape_a).astype("float32")
data_b = np.random.uniform(-1, 1, size=shape_b).astype("float32")
matmul_0 = np.matmul(data_a, data_b)
orig_k = 2016
k_num = 8
k_tile = orig_k // k_num
data_a1 = data_a.reshape([1, 49, k_num, k_tile])
data_a2 = np.transpose(data_a1, [0, 2, 1, 3])
data_b1 = data_b.reshape([k_num, k_tile, 448])
matmul_1 = np.matmul(data_a2, data_b1)
matmul_2 = np.sum(matmul_1, axis=-3)
error = matmul_0 - matmul_2