Getting started with rWCVP
introduce
The World Catalog of Vascular Plants (WCVP) is a global consensus of vascular plant species. It provides the names, synonyms, taxonomy and distribution of >340,000 vascular plant species known to science.
The authors developed rWCVP to make some common tasks using WCVP easier. These include standardizing lists of taxon names according to WCVP, obtaining and mapping species distributions, and creating inventories of taxa found in specific regions.
1. Access to WCVP
For the functions in rWCVP to work properly, you need to have access to a copy of WCVP.
1.1 Method 1
One way to load WCVP is to install the associated data package rWCVPdata:
if (!require(rWCVPdata)) {
install.packages("rWCVPdata",
repos = c(
"https://matildabrown.github.io/drat",
"https://cloud.r-project.org"
)
)
}
== Wait for the download and installation to complete, == You can see the taxa and distribution data of rWCVPdata :
library(rWCVPdata)
names = rWCVPdata::wcvp_names
distribution = rWCVPdata::wcvp_distributions
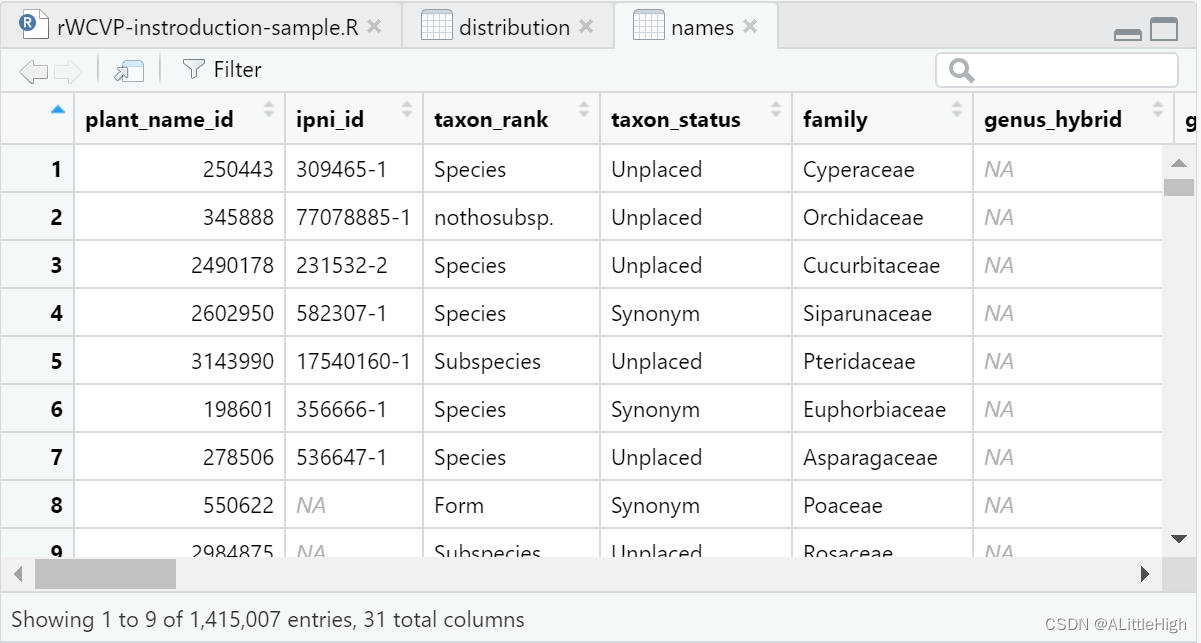
rWCVPdata contains more than 1.4 million species information:
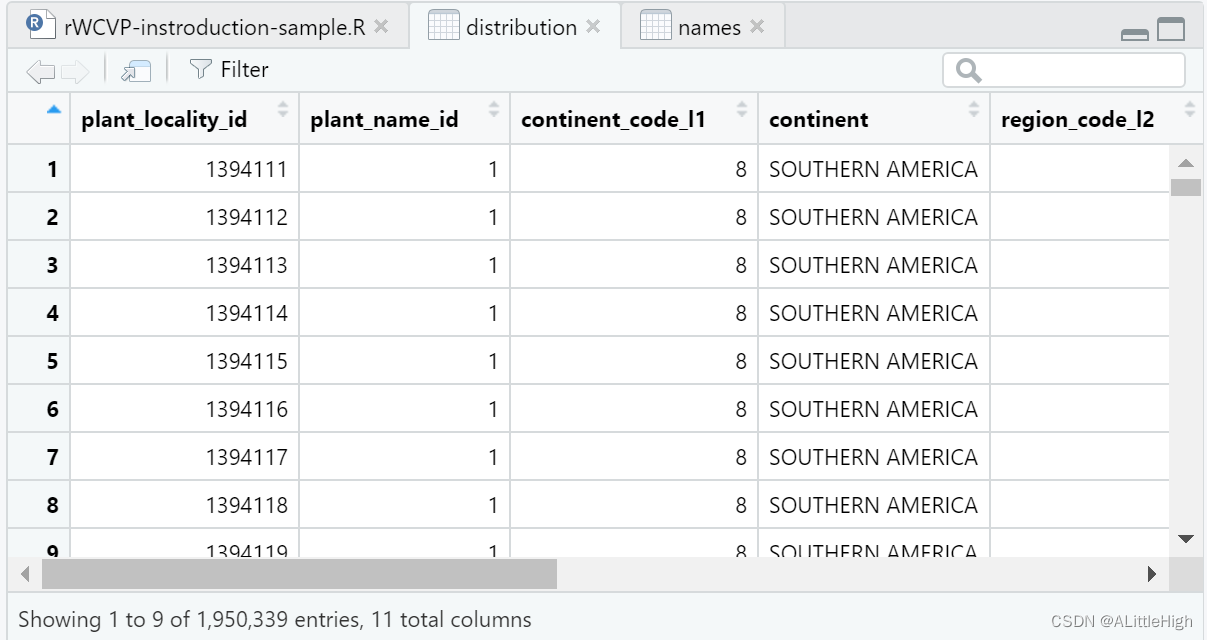
rWCVPdata contains more than 1.9 million species distribution information:
1.2 Method 2 (cautious)
If the data package is not available, or you wish to use a different version of WCVP, you can provide a local copy of the data to the main function in this package. For example, to generate a manifest:
names <- read_csv("/path/to/wcvp_names.csv")
distributions <- read_csv("/path/to/wcvp_distributions.csv")
checklist <- wcvp_checklist("Acacia",
taxon_rank = "genus", area_codes = "CPP",
wcvp_names = names, wcvp_distributions = distributions
)
Be careful if you are using your own version of WCVP! The structure of the WCVP table sometimes varies from version to version. rWCVP should be set up to work with the latest version of WCVP and any previous versions that share the same structure.
2. WCVP data screening
Some rWCVP functions involve WCVP data screening to generate lists or summaries of vascular plant species in specific regions.
These functions accept two parameters for filtering WCVP:
- taxon : The name of a valid taxon with taxonomic rank of species or higher (e.g. species "Myrcia almasensis", genus "Myrcia" or "Myrtaceae" families).
- area : A vector of WGSRPD Level 3 codes for the area to focus on.
These parameters can be used in combination in wcvp_checklist, wcvp_occ_mat and wcvp_summary to produce an output of focal taxa in the desired region. For example, the Brazilian myrtaceae :
# filter Myrtaceae species in Brazil
checklist <- wcvp_checklist("Myrtaceae",
taxon_rank = "family",
area_codes = c("BZC", "BZN", "BZS", "BZE", "BZL")
)

2.1 Notes on filtering by taxon
When filtering by taxon, you need to use the taxon.rank parameter to tell the function the taxon rank of the name you provide
For example, to generate a summary table for Poa annua:
# summary table for Poa
wcvp_summary("Poa", taxon_rank = "genus")
ℹ No area specified. Generating global summary.
$Taxon
[1] "Poa"
$Area
[1] "the world"
$Grouping_variable
[1] "area_code_l3"
$Total_number_of_species
[1] 573
$Number_of_regionally_endemic_species
[1] 573
$Summary
# A tibble: 280 × 6
area_code_l3 Native Endemic Introduced Extinct Total
<chr> <int> <int> <int> <int> <int>
1 ABT 21 0 5 0 26
2 AFG 23 1 0 0 23
3 AGE 13 2 5 0 18
4 AGS 24 0 10 0 34
5 AGW 34 8 3 0 37
6 ALA 6 0 3 0 9
7 ALB 17 0 0 0 17
8 ALG 8 0 1 0 11
9 ALT 30 3 1 0 31
10 ALU 7 0 3 0 10
# ℹ 270 more rows
# ℹ Use `print(n = ...)` to see more rows
You can provide the taxon name of the rank Species, Genus, Family, Order or higher. WCVP only provides classification information below the department level. We have included a table called taxonomic_mapping for mapping families to orders and higher taxonomies according to APG IV.
# APG Ⅳ
head(taxonomic_mapping)
higher order family
1 Angiosperms Acorales Acoraceae
2 Angiosperms Alismatales Alismataceae
3 Angiosperms Alismatales Aponogetonaceae
4 Angiosperms Alismatales Araceae
5 Angiosperms Alismatales Butomaceae
6 Angiosperms Alismatales Cymodoceaceae
We use this table behind the scenes to filter using these higher category ranks. For example, to aggregate across all Gramineae :
# summary all Poales:
wcvp_summary("Poales", taxon_rank = "order")
ℹ No area specified. Generating global summary.
$Taxon
[1] "Poales"
$Area
[1] "the world"
$Grouping_variable
[1] "area_code_l3"
$Total_number_of_species
[1] 23770
$Number_of_regionally_endemic_species
[1] 23770
$Summary
# A tibble: 368 × 6
area_code_l3 Native Endemic Introduced Extinct Total
<chr> <int> <int> <int> <int> <int>
1 ABT 359 0 70 0 429
2 AFG 485 16 15 0 504
3 AGE 908 31 166 0 1074
4 AGS 389 20 88 0 477
5 AGW 890 130 105 0 995
6 ALA 711 2 153 0 864
7 ALB 363 4 14 0 387
8 ALD 44 7 13 0 57
9 ALG 445 11 41 0 497
10 ALT 383 9 6 0 389
# ℹ 358 more rows
# ℹ Use `print(n = ...)` to see more rows
2.2 Explanation on screening by distribution area
WCVP lists taxon distributions at level 3 using the World Geographic Scheme for Recorded Plant Distributions (WGSRPD) . This level corresponds to "plant states", which mostly follow state borders, unless large states are divided or outlying areas are omitted. Functions in rWCVP expect area to be provided as a vector of WGSRPD level 3 code.
Finding your entire area of interest can be annoying. For example, to filter by species in Brazil, you would provide a vector of 5 codes. For convenience, rWCVP has a function to convert region names to WGSRPD Level 3 code vectors.
# convert region to codes
get_wgsrpd3_codes("Brazil")
ℹ Matches to input geography found at Country (Gallagher) and Region (Level 2)
[1] "BZC" "BZE" "BZL" "BZN" "BZS"
This can be fed directly into the function that filters WCVP by region.
wcvp_summary("Poa", taxon_rank = "genus", area = get_wgsrpd3_codes("Southern Hemisphere"))
ℹ Matches to input geography found at Hemisphere level
ℹ Including WGSRPD areas that span the equator. To turn this off, use `include_equatorial = FALSE`
$Taxon
[1] "Poa"
$Area
[1] "Southern Hemisphere (incl. equatorial Level 3 areas)"
$Grouping_variable
[1] "area_code_l3"
$Total_number_of_species
[1] 264
$Number_of_regionally_endemic_species
[1] 237
$Summary
# A tibble: 74 × 6
area_code_l3 Native Endemic Introduced Extinct Total
<chr> <int> <int> <int> <int> <int>
1 AGE 13 2 5 0 18
2 AGS 24 0 10 0 34
3 AGW 34 8 3 0 37
4 ANT 0 0 1 0 1
5 ASC 0 0 1 0 1
6 ASP 2 1 3 0 5
7 ATP 8 1 3 0 11
8 BOL 31 2 3 0 34
9 BOR 2 1 0 0 2
10 BUR 3 0 0 0 3
# ℹ 64 more rows
# ℹ Use `print(n = ...)` to see more rows
author practice
m = wcvp_checklist(taxon = "Mahonia", taxon_rank = "genus")
b = wcvp_checklist(taxon = "Berberis", taxon_rank = "genus")

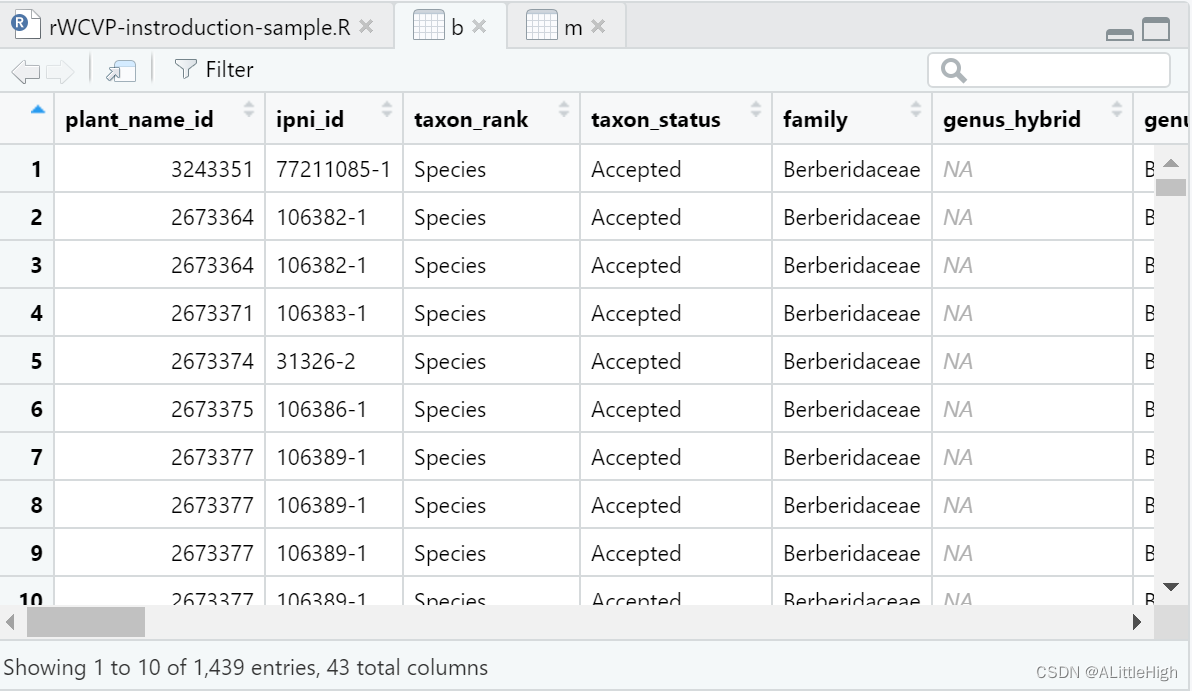
m means mahonia, b means berberis. In WCVP, the independent status of mahonia is not recognized, and it is incorporated into berberis. Maybe get berberis from rWCVP, and then try to pick Mahonia from U.Taxonstand.