In recent years, with the development of high-throughput sequencing technology, transcriptome sequencing has become the main means to study the regulation of gene expression. We know that the transcripts of many species are very diverse and complex, and most eukaryotic genes do not conform to the "one gene, one transcript" model, and these genes often have multiple spliced forms. Through next-generation sequencing, gene expression and quantitative research can be carried out very accurately, but due to the limitation of read length, the information of full-length transcripts cannot be obtained. Therefore, the full-length transcriptome based on the third-generation sequencing platform has become a new research boom.
Full-length transcriptome (Full-length transcriptome) is based on PacBio and Nanopore three-generation sequencing platform, without interrupting the splicing, directly obtains the full-length mRNA sequence and complete structure information including 5'UTR, 3'UTR, and polyA tail, so as to accurately analyze There are structural information such as alternative splicing and fusion genes of species with reference genomes, which overcomes the problems of short transcript splicing and incomplete information of species without reference genomes. At the same time, with the help of next-generation sequencing data, transcript-specific expression analysis can be performed to obtain more comprehensive annotation information.
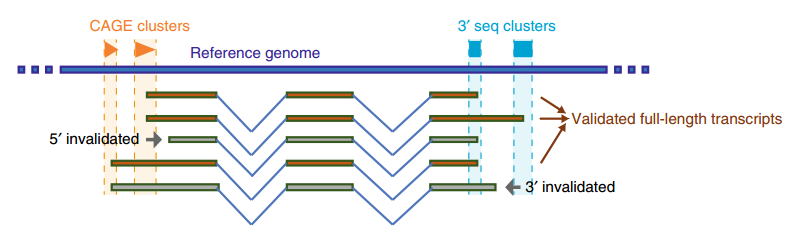
Figure 1 Confirmed full-length transcripts in rats
Principles of Full-Length Transcriptome Sequencing
experiment process
Shi YanLiuCh

Figure 2 Full-length transcriptome experiment process
CCS Sequencing (Rolling Circle Sequencing)
GunHuanCeXu
The full-length transcriptome uses single-molecule real-time sequencing technology to construct a dumbbell-shaped library and cycle-sequence it in a circular manner. Short-fragment libraries are more likely to fall into zero-mode waveguides (ZMWs) during sequencing.
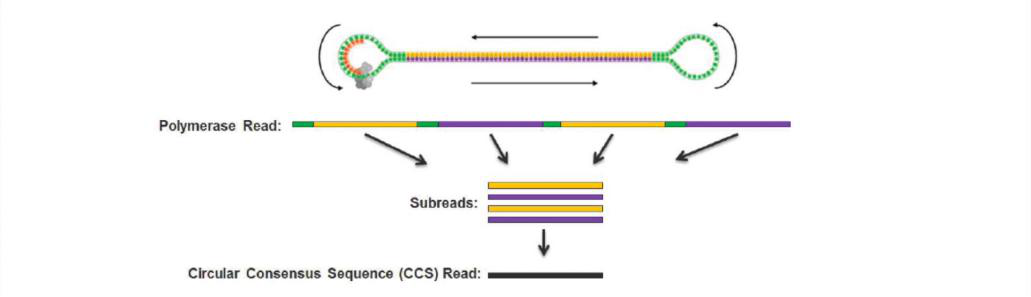
Figure 3 Full-length transcriptome CCS sequencing
-
Polymerase read: The sequence directly read by the third-generation sequencing, including adapters and inserts;
-
Subreads: Remove the Polymerase read adapter, and generate a subread every time the insert fragment is tested;
-
CCS read: The consensus sequence generated by subreads (at least two) derived from the same Polymerase read.
Full-length transcription project design
For species with reference genomes , full-length transcriptome information can correct genome misassembly, more accurately discover new transcripts and genes, and analyze gene fusion events.
For species with incomplete reference genomes , the full-length transcriptome can optimize the gene structure, assist genome assembly and annotation, and improve the accuracy of gene expression and data utilization.
For species without a reference genome , the species Unigene library is constructed by three-generation full-length transcriptome sequencing, and the reference sequence at the transcriptome level of the species (reference genome at the transcriptome level) can be obtained without sequence assembly, providing a great deal of support for subsequent research. Good genetic information base.
The transcriptome has spatiotemporal specificity, and the experimental design is carried out according to the research purpose, as follows:
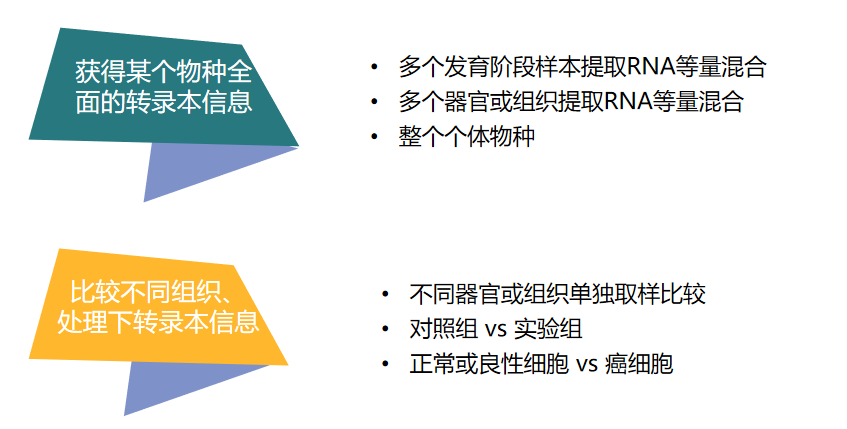
Figure 4 Full-length transcriptome experimental design

It is recommended to use 2+3 generations of transcriptome sequencing technology at the same time to ensure structural accuracy, sequence integrity and sequence expression accuracy, to achieve the optimal use of data and the highest cost performance. According to different requirements and purposes, for most species, the recommended amount of sequencing data is 8-12G. To study low-expressed genes, variable splicing events, polyploids, or species with a large number of genes, it is necessary to increase the amount of data appropriately.
Full-length transcriptome analysis workflow
After obtaining the original sequencing sequence (Sequenced Reads), the bioinformatics analysis is carried out through the following process:
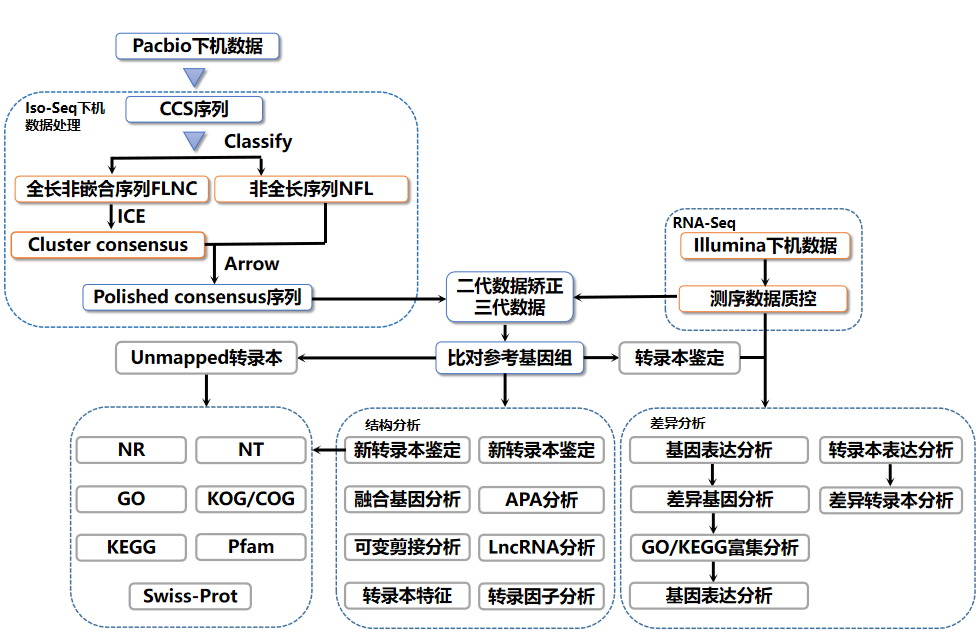
Figure 5 Bioinformatics analysis process of the full-length transcriptome of Lingen Biotech
Advantages of Lingen Bio's full-length transcriptome sequencing
-
Without the need for transcript splicing, the full-length transcript sequence can be directly obtained, thereby more truly reflecting the transcriptome information of the sequenced species;
-
Able to detect multiple alternative splicing forms, discover more splice sites and alternative splicing events;
-
Ability to discover new functional genes and supplement genome annotations;
-
Contributes to the precise analysis of fusion genes, homologous genes, superfamily genes or alleles;
-
Professional bioinformatics analysis team with rich project experience.
Sample request
-
Total RNA Sample Requirements
Generally, total RNA>5 ug/library, total amount>15 ug (3 libraries), concentration ≧250ng/ul.
-
RNA sample quality requirements
OD260/280≧1.8, OD260/230≧1.0, normal peak at 260nm; RIN value of total RNA≧8.0, 28S/18S≧1.3.
-
avoid cDNA damage
Safe dyes are used to avoid damage to cDNA caused by EB and ultraviolet radiation.
In short , the three-generation full-length transcriptome is applicable to both species with ginseng and species without ginseng, and more transcriptional regulatory events can be obtained, thereby more truly reflecting the transcriptome information of the sequenced species.
At present , full-length transcriptome sequencing to study gene structure has become a trend of publishing articles. The research species include sorghum, corn, Arabidopsis, chicken, human and mouse, bamboo, cotton, etc. Lingen Biotech provides professional full-length transcriptome sequencing and analysis services to assist your research!

Do a good job of sequencing with heart and entrust every result