Introduction
- Sequence Read Archive (SRA) data is available through multiple cloud providers and NCBI servers. It is the largest publicly available high-throughput sequencing data repository.
- SRA accepts data from various departments of life, metagenomics and environmental surveys.
- SRA stores raw sequencing data and comparison information to improve reproducibility and facilitate new discoveries through data analysis.
- The NCBI website stores the database of the raw data of second-generation sequencing
- SRA official website
Download SRA sequence from Entrez search results
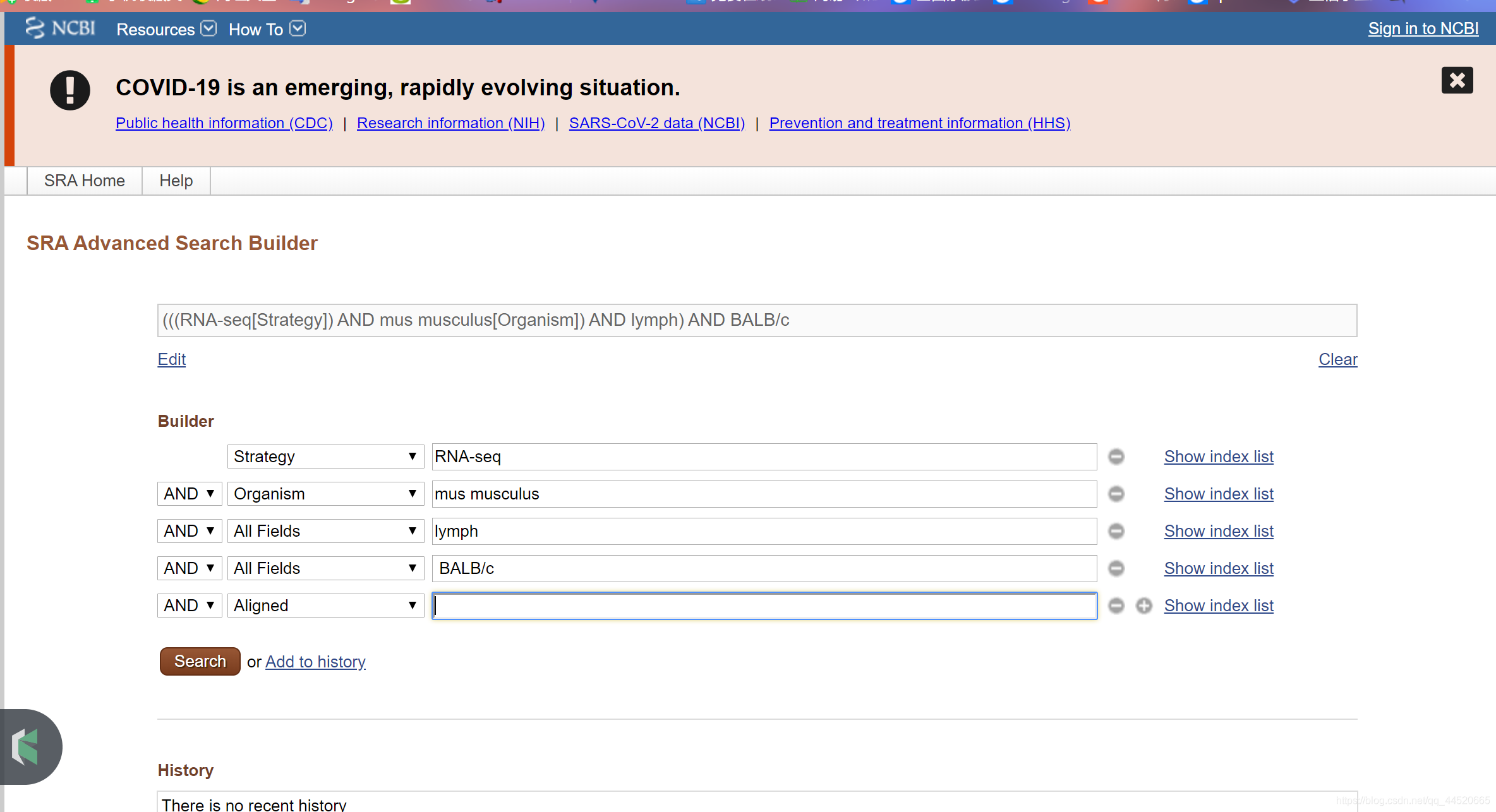
For example, find RNA-Seq records of BALB/c mouse lymph node tissue in SRA Entrez
Get search results
- Advanced search in SRA search: (((“mus musculus”[Organism]) AND BALB/c*) AND “lymph*”) AND “rna seq”[Strategy]
- To limit your search to aligned data only, add AND alignment data [attribute] to the query above.
- Click the check box next to the record (experiment) to select the data of interest. Uncheck all check boxes to select all records (experiments) from the search.
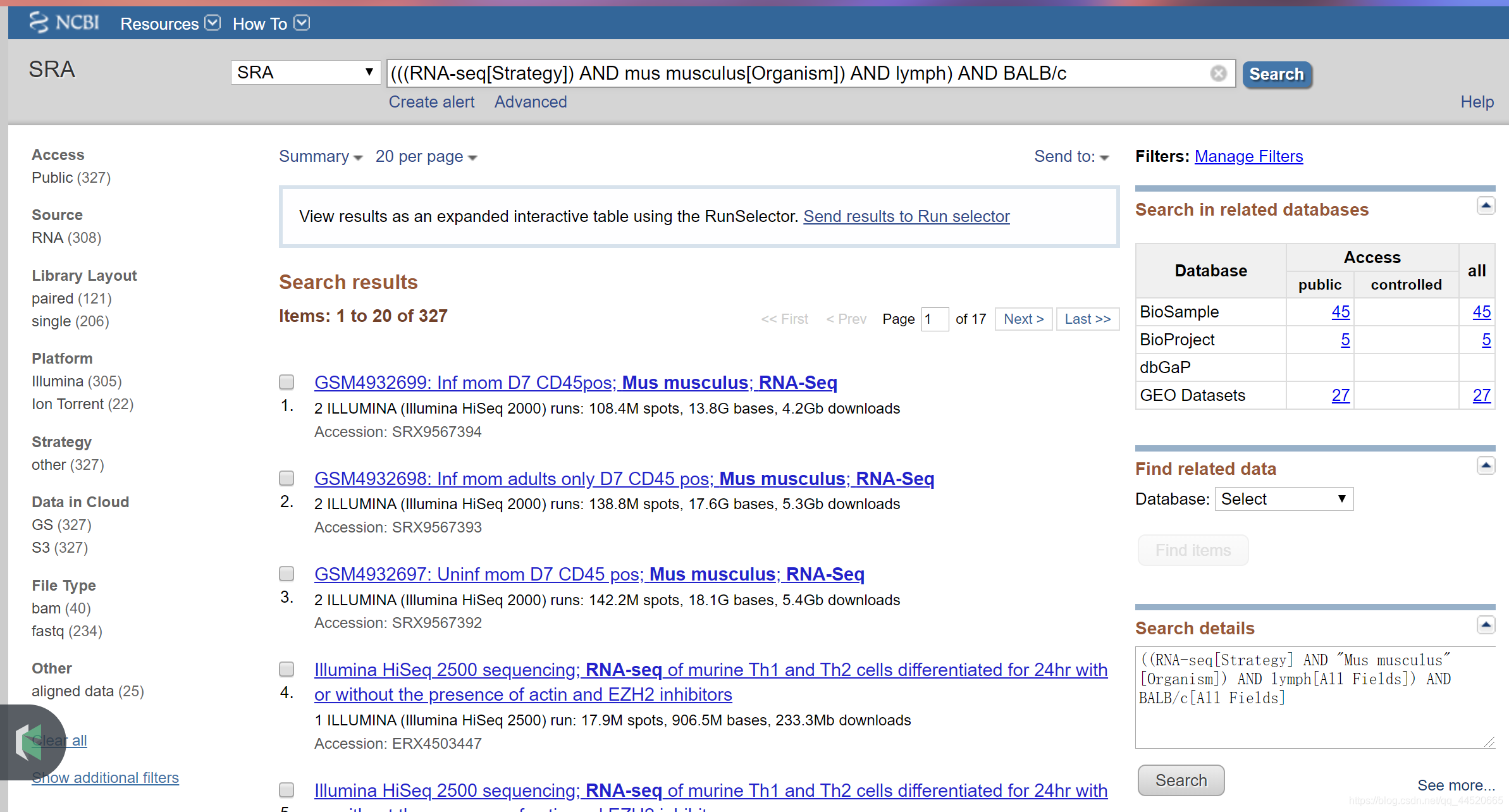
Get run accessions
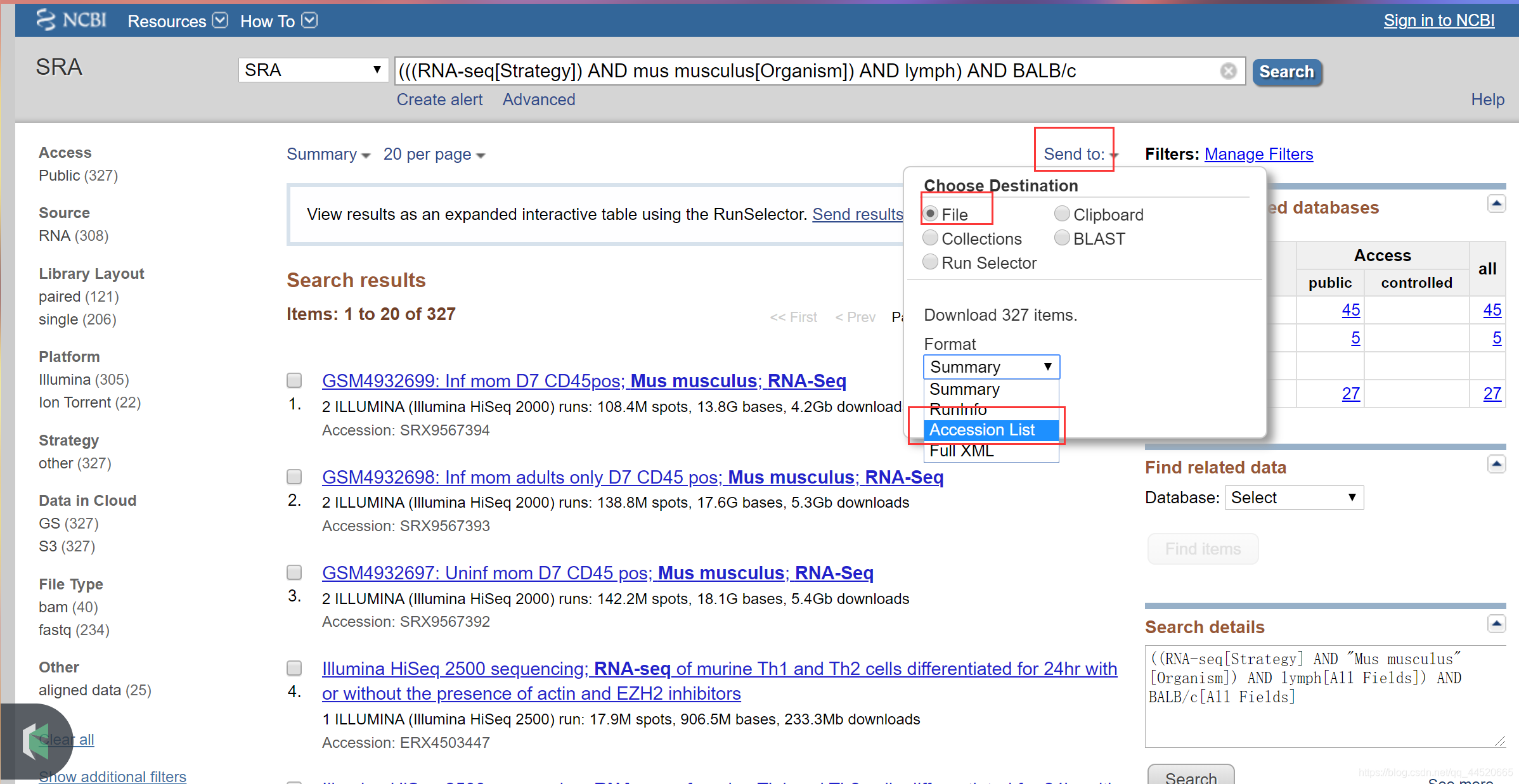
Run accessions are used to download SRA data. To download the list of run accessions selected in your Entrez search, do the following:
- Click send to at the top of the page, select the radio button File, and select Accession List
- Save this file in the location where you run the SRA toolkit.
The format of the Sraacclist.txt file is as follows:
SRR11192680
SRR11192681
SRR11192682
SRR11192683
SRR11192684
Use the SRA toolkit to download sequence data files
- The SRA run file contains only sequence data and does not contain any metadata linked to the run (sample information, etc.)
- Please make sure you are running the latest version of the toolkit, because the earlier version may not be compatible with the latest loaded data or the latest network protocol
Install SRA Toolkit:
- Download sratoolkit.current-win64.zip: http://ftp-trace.ncbi.nlm.nih.gov/sra/sdk/current/sratoolkit.current-win64.zip
- path:C:\Users\xuyunfeng\Desktop\sratoolkit.current-win64\sratoolkit.2.10.9-win64
- Open a command shell, such as start or run == cmd.exe==
- cdGo to the directory where you unzipped the zip file
- cd bin

Configure SRA Toolkit
- Only a small number of options need to be enabled to access public and controlled access data in the cloud. To start the configuration, run:vdb-config -i
- You will see a screen where you can manipulate the buttons by pressing the letter highlighted in red, or pressing the Tab key until you reach the desired button, and then pressing the space bar or the Enter key.
- You want to enable the "Remote access" option on the main screen.
- Go to the "Cache" tab, where you will enable "Local File Cache" and set the "Location of User Repository".
- The repository directory needs to be set to an empty folder. This is the folder where the prefetch will store the files.
- Go to your cloud provider tab and accept "Report Cloud Instance Identity"
- The cloud instance identity only reports the cloud you are using (AWS v GCP), so you can access the data for free.
Check if the toolkit is available
fastq-dump --stdout -X 2 SRR390728
After a few seconds, the command generates the following output
Read 2 spots for SRR390728
Written 2 spots for SRR390728
@SRR390728.1 1 length=72
CATTCTTCACGTAGTTCTCGAGCCTTGGTTTTCAGCGATGGAGAATGACTTTGACAAGCTGAGAGAAGNTNC
+SRR390728.1 1 length=72
;;;;;;;;;;;;;;;;;;;;;;;;;;;9;;665142;;;;;;;;;;;;;;;;;;;;;;;;;;;;;96&&&&(
@SRR390728.2 2 length=72
AAGTAGGTCTCGTCTGTGTTTTCTACGAGCTTGTGTTCCAGCTGACCCACTCCCTGGGTGGGGGGACTGGGT
+SRR390728.2 2 length=72
;;;;;;;;;;;;;;;;;4;;;;3;393.1+4&&5&&;;;;;;;;;;;;;;;;;;;;;<9;<;;;;;464262
Download public data
- PrefetchIt is part of the SRA toolkit. This program downloads Runs (compressed sequence files in SRA format) and all other data needed to convert Run from SRA format to a more common format. Prefetch can be used to correct and complete incomplete Run downloads
- Use this prefetch command to download Run from the previous example in SRA format
$ prefetch SRR000001
List of Runs:
prefetch --option-file SraAccList.txt
- fastq-dump和sam-dumpIt is also part of the SRA toolkit and can be used to convert pre-extracted runs from compressed SRA format to fastq or sam format , for example:
fasterq-dump --split-files SRR11180057.sra
- You can also avoid the prefetching step by entering only Run accession without the .sra extension in the fastqq-dump or sam-dump command and download and convert the operation step by step:
fasterq-dump --split-files SRR11180057
Download the original submitted file
- If you wish to use the original submitted file instead of dumping standardized data from the archive, SRA has stored the original submitted file in a cloud storage bucket that can be accessed through the prefetch command.
- See Download SRA sequence data using Amazon Web Services (AWS)
- For example, the prefetch command
prefetch --type fastq SRR11180057
Use ==–type== command to specify the type of file to be downloaded. You can find the file type of the original file in the " Data Access " tab in BigQuery's SRA or "Run Browser" , or use any to get all available formats.
Download protected data
For information on how to download dbGaP data, please see: Protected Data Usage Guide
Download metadata related to SRA data
From the search results page
- The SRA Run file does not contain any information about metadata linked to the data itself (example information, etc.).
- To download the metadata of each Run in the Entrez query, click Send to at the top of the page, select the File radio button, and then select RunInfo from the drop-down menu.
- This will generate a tabularSraRunInfo.csvFile containing metadata available for each run.
From Run Selector
From Run Selector download a slightly different set of metadata in a tab-delimited file
to download the metadata for each Run Entrez query, do the following:
- Click Send to the top of the page, check the Run option radio button, and then click the button to enter.
- If necessary, use the various filters provided in the Run Selector interface to refine the results.
- Click the "Run Information Sheet" button. This will generate a tableSraRunTable.txtFile containing metadata available for each run.
Download sequence data from Run Browser
Run Browser allows limited downloading of HTTP misaligned and aligned sequences
Examples of unaligned sequences
- Open the selected run in Run Browser .
- Click the Reads tab.
By applying Filter to find some reading material or filter criteria field empty.
Click the Filtered Download button.
Select the available download format and click the Download link.
Aligned sequence example
Open the selected run in Run Browser .
Click the Alignment tab.
Select the available download format in the drop-down menu, and then click the Screen or File button to output the run to the screen or file.