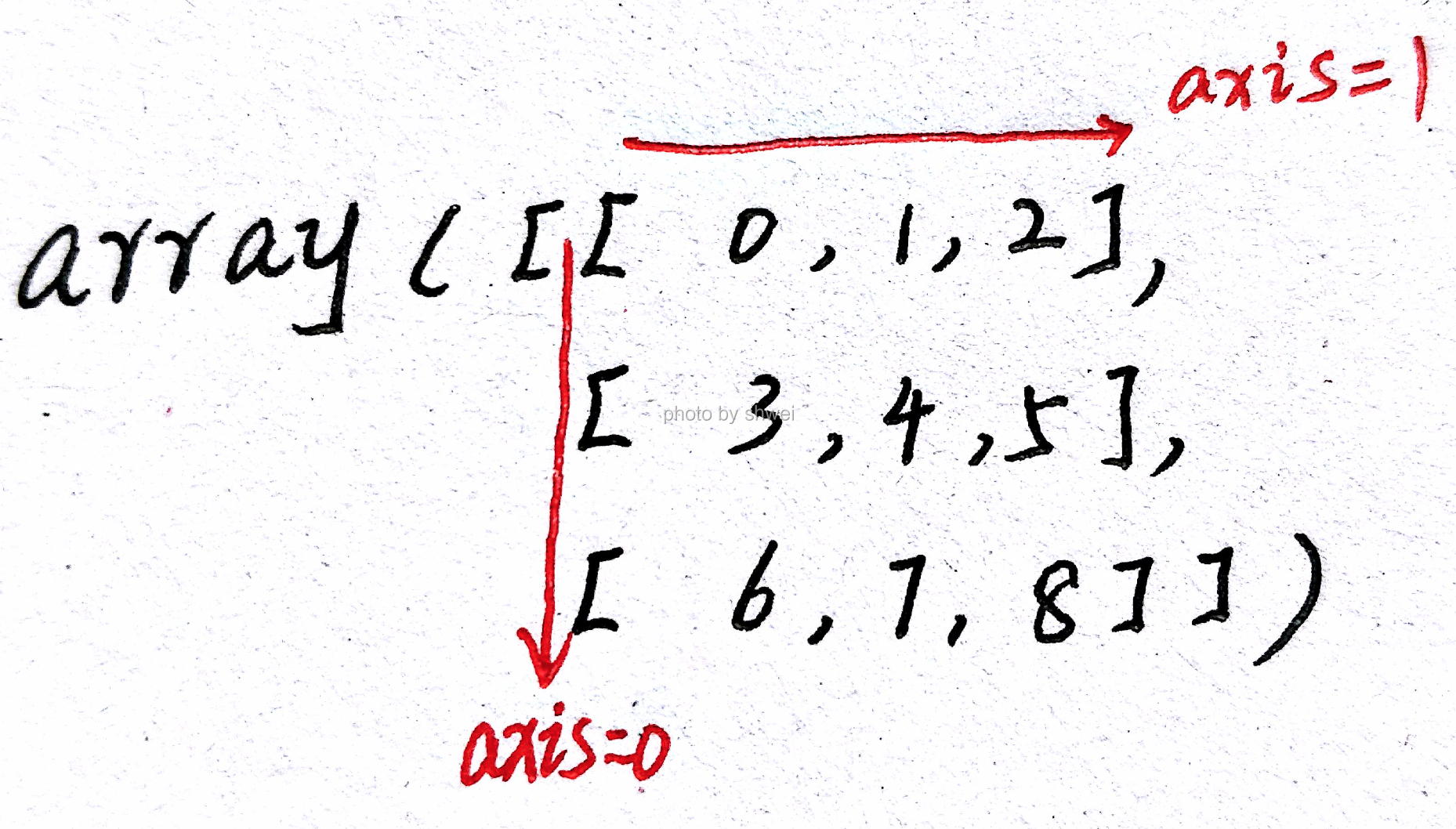文章目录
- 4.1 NumPy ndarray
- 4.1.1 Create ndarray
- 4.1.2 Data Types for ndarrays
- 4.1.3 Operations between Arrays and Scalars
- 4.1.4 Basic Indexing and Slicing
- 4.1.5 Boolean Indexing
- 4.1.6 Fancy Indexing
- 4.1.7 Transposing Arrays and Swapping Axes
- 4.2 Universal Functions: Fast Element-wise Array Functions
- 4.3 Data Processing Using Arrays
- 4.3.0 np.meshgrid()
- 4.3.1 Expressing Conditional Logic as Array Operations
- 4.3.2 Mathematical and Statistical Methods
- 4.3.3 Methods for Boolean Arrays
- 4.3.4 Sorting
- 4.3.5 Unique and Other Set Logic
- 4.4 File Input and Output with Arrays
- 4.5 Linear Algebra
- 4.6 Random Number Generation
- NumPy Summay
4.1 NumPy ndarray
NumPy, short for Numerical Python, is the fundamental package required for high performance scientific computing and data analysis.
One of the key features of NumPy is its N-dimensional array object, or ndarray, which
is a fast, flexible container for large data sets in Python.
Whenever we see “array”, “Numpy array”, or “ndarray” in this book, with few exceptions they all refer to the same thing: the ndarray object
A ndarray example
array([[7, 5],
[2, 3]])
import numpy as np
my_arr = np.arange(1000000)
my_list = list(range(1000000))
Run time
%time my_arr2 = my_arr * 2
%time my_list = my_list * 2
CPU times: user 3.99 ms, sys: 5.66 ms, total: 9.64 ms
Wall time: 13.1 ms
CPU times: user 20.3 ms, sys: 9.21 ms, total: 29.5 ms
Wall time: 39.1 ms
As we can see Numpy method is much faster than Python method, and cost less space.
4.1.1 Create ndarray
The easiest way to create an array is to use the array function
# data1 is a list
data1 = [[1, 2, 3], [4, 5, 6]]
array1 = np.array(data1)
array1
array1.shape
(2, 3)
data2 = [5, 4.1, 2, 8]
array2 = np.array(data2)
print(array2)
print(array2.dtype)
[5. 4.1 2. 8. ]
float64
print(np.zeros(4))
[0. 0. 0. 0.]
To create a higher dimensional array with these methods, pass a tuple for the shape
np.zeros((2,5)) # parameter Shape: int or tuple int
array([[0., 0., 0., 0., 0.],
[0., 0., 0., 0., 0.]])
np.ones((3,4))
array([[1., 1., 1., 1.],
[1., 1., 1., 1.],
[1., 1., 1., 1.]])
empty creates an array without initializing its values to any particular value:
np.empty((1,2,3))
array([[[1.28822975e-231, 1.28822975e-231, 1.97626258e-323],
[0.00000000e+000, 0.00000000e+000, 0.00000000e+000]]])
arange is an array-valued version of the built-in Python range function:
np.arange(1, 10, 2)
array([1, 3, 5, 7, 9])
list(range(1, 10, 2)) # function range return an iterable object, not a list
[1, 3, 5, 7, 9]
ones_like takes another array and produces a ones array of the same shape and dtype
np.ones_like([2,3,4])
array([1, 1, 1])
It will tell us the right parameters format, so we don’t need to memory them.
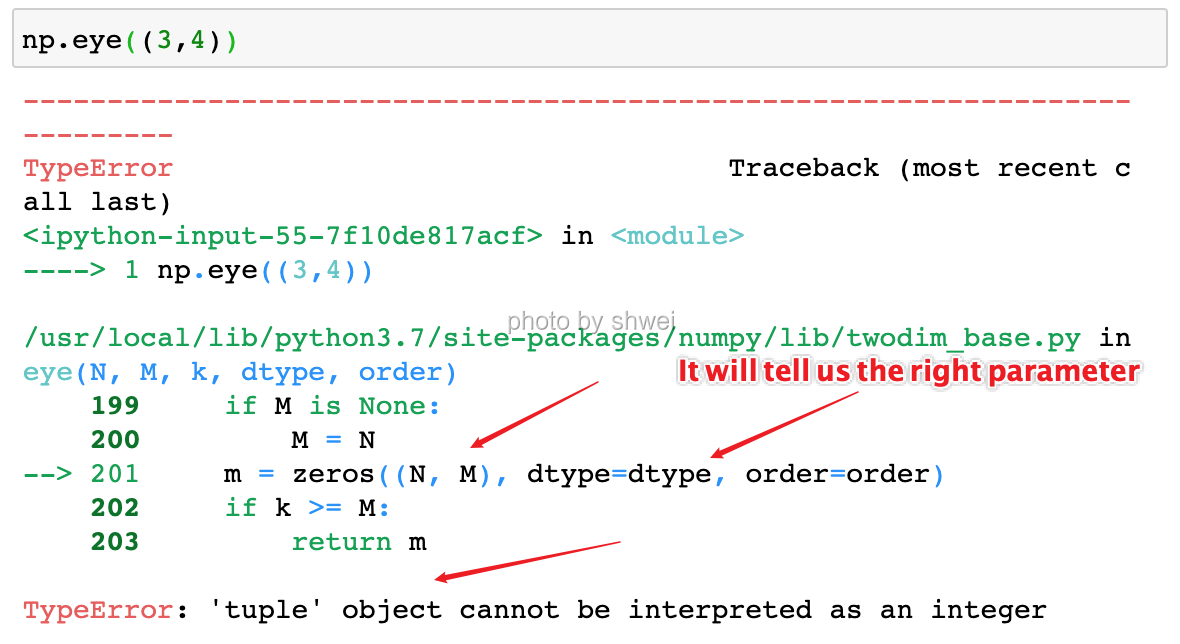
Create a square N * N identity matrix (1’s on the digonal and 0’s elsewhere)
np.identity(4)
array([[1., 0., 0., 0.],
[0., 1., 0., 0.],
[0., 0., 1., 0.],
[0., 0., 0., 1.]])
np.eye(3,4)
array([[1., 0., 0., 0.],
[0., 1., 0., 0.],
[0., 0., 1., 0.]])
np.full([2,3], 5)
array([[5, 5, 5],
[5, 5, 5]])
Table 4-1. Array creation functions
| Function | Description |
|---|---|
| array | Convert input data (list, tuple, array, or other sequence type) to an ndarray either by inferring a dtype or explicitly specifying a dtype. Copies the input data by default. |
| asarray | Convert input to ndarray, but do not copy if the input is already an ndarray |
| arange | Like the built-in range but returns an ndarray instead of a list. |
| ones, ones_like | Produce an array of all 1’s with the given shape and dtype. ones_like takes another array and produces a ones array of the same shape and dtype. |
| zeros, zeros_like | Like ones and ones_like but producing arrays of 0’s instead |
| empty, empty_like | Create new arrays by allocating new memory, but do not populate with any values like ones and zeros |
| eye, identity | Create a square N x N identity matrix (1’s on the diagonal and 0’s elsewhere) |
4.1.2 Data Types for ndarrays
The data type or dtype is a special object containing the information the ndarray needs to interpret a chunk of memory as a particular type of data:
The default dtype is float64
arr1 = np.array([1,2,3], dtype=np.float64)
arr2 = np.array([1,2,3.1], dtype=np.int32)
arr3 = np.ones(4) # use default dtype
arr1.dtype
dtype('float64')
arr1
array([1., 2., 3.])
arr2.dtype
dtype('int32')
arr2
array([1, 2, 3], dtype=int32)
arr3.dtype
dtype('float64')
arr3
array([1., 1., 1., 1.])
Table 4-2. NumPy data types
| Type | Type Code | Description |
|---|---|---|
| int8, uint8 | i1, u1 | Signed and unsigned 8-bit (1 byte) integer types |
| int16, uint16 | i2, u2 | Signed and unsigned 16-bit integer types |
| int32, uint32 | i4, u4 | Signed and unsigned 32-bit integer types |
| int64, uint64 | i8, u8 S | igned and unsigned 32-bit integer types |
| float16 | f2 | Half-precision floating point |
| float32 | f4 or f | Standard single-precision floating point. Compatible with C float |
| float64, float128 | f8 or d | Standard double-precision floating point. Compatible with C double and Python float object |
| float128 | f16 or g | Extended-precision floating point |
| complex64, complex128, complex256 | c8, c16, c32 | Complex numbers represented by two 32, 64, or 128 floats, respectively |
| bool | ? | Boolean type storing True and False values |
| object | O | Python object type |
| string_ | S | Fixed-length string type (1 byte per character) . For example, to create a string dtype with length 10, use ‘S10’. |
| unicode_ | U | Fixed-length unicode type (number of bytes platform specific). Same specification semantics as string_ (e.g. ‘U10’). |
4.1.3 Operations between Arrays and Scalars
Arrays are important because they enable you to express batch operations on data without any for loops. This is usually called vectorization.
arr = np.array([[1,2,3],[4.,5,6]])
arr
array([[1., 2., 3.],
[4., 5., 6.]])
arr * arr
array([[ 1., 4., 9.],
[16., 25., 36.]])
1 / arr
array([[1. , 0.5 , 0.33333333],
[0.25 , 0.2 , 0.16666667]])
arr ** 0.5
array([[1. , 1.41421356, 1.73205081],
[2. , 2.23606798, 2.44948974]])
arr2 = np.array([[0., 4., 1.], [7, 2,12.]])
arr2
array([[ 0., 4., 1.],
[ 7., 2., 12.]])
Comparing between two arrays of the same size return bool array
arr2 > arr
array([[False, True, False],
[ True, False, True]])
Operations between differently sized arrays is called broadcasting. Having a deep understanding of broadcasting is not neccessay for most of this book.
4.1.4 Basic Indexing and Slicing
An important first distinction frome lists is that array slice are views on the original array. This means that the data is not copied, and any motifications to the view will be reflected in the source array.
arr = np.arange(10)
arr
array([0, 1, 2, 3, 4, 5, 6, 7, 8, 9])
# any motifications to the view will be reflected in the source array:
arr[5:8] = 12
arr
array([ 0, 1, 2, 3, 4, 12, 12, 12, 8, 9])
# array slice is not copied
arr_slice = arr[5:8]
arr_slice[1] = 100
arr
array([ 0, 1, 2, 3, 4, 12, 100, 12, 8, 9])
# list slice is copied
list_ = list(range(10))
list_slice = list_[5:8]
list_slice[1] = 100
list_
[0, 1, 2, 3, 4, 5, 6, 7, 8, 9]
Notes: As Numpy has been designed with large data use cases in mind, you could imagine performance and memory problems if Numpy insisted on copying data left and right.
# array slice is copied if you use copy()
arr_slice = arr[5:8].copy()
arr_slice[1] = 888
arr
array([ 0, 1, 2, 3, 4, 12, 100, 12, 8, 9])
higher dimensional array index
arr2d = np.array([[1,2],[3,4]])
arr2d[0]
array([1, 2])
You can pass a comma-separated list of indices to select individual elements.
So these are equivalent:
arr2d[0][1]
2
arr2d[0,1]
2
Note: they are different if it is a slice
4.1.4.1 Note the function of arr3d[ :2, 1:, 0]
use comma , to process each dimension
arr2d[:2,:1] # process each dimension
array([[1],
[3]])
arr2d[:2][:1]
# equivalent: tmp = arr2d[:2] and tmp[:1]
array([[1, 2]])
3D array
arr3d = np.array([ [[1,2,3], [4,5,6]],
[[7,8,9], [10,11,12]]
])
arr3d
array([[[ 1, 2, 3],
[ 4, 5, 6]],
[[ 7, 8, 9],
[10, 11, 12]]])
integer indexes return lower dimensonal axes
arr3d[0]
array([[1, 2, 3],
[4, 5, 6]])
colon: return the same dimensional axes
arr3d[:1]
array([[[1, 2, 3],
[4, 5, 6]]])
arr3d[0,1]
array([4, 5, 6])
arr3d[0,1] = 0
arr3d
array([[[ 1, 2, 3],
[ 0, 0, 0]],
[[ 7, 8, 9],
[10, 11, 12]]])
Note that in all of these cases where subsections of the array have been selected, the returned arrays are views, not copied.
Indexing with slices
arr2d = np.array([[1,2,3],
[4,5,6],
[7,8,9]
])
arr2d
array([[1, 2, 3],
[4, 5, 6],
[7, 8, 9]])
arr2d.shape
(3, 3)
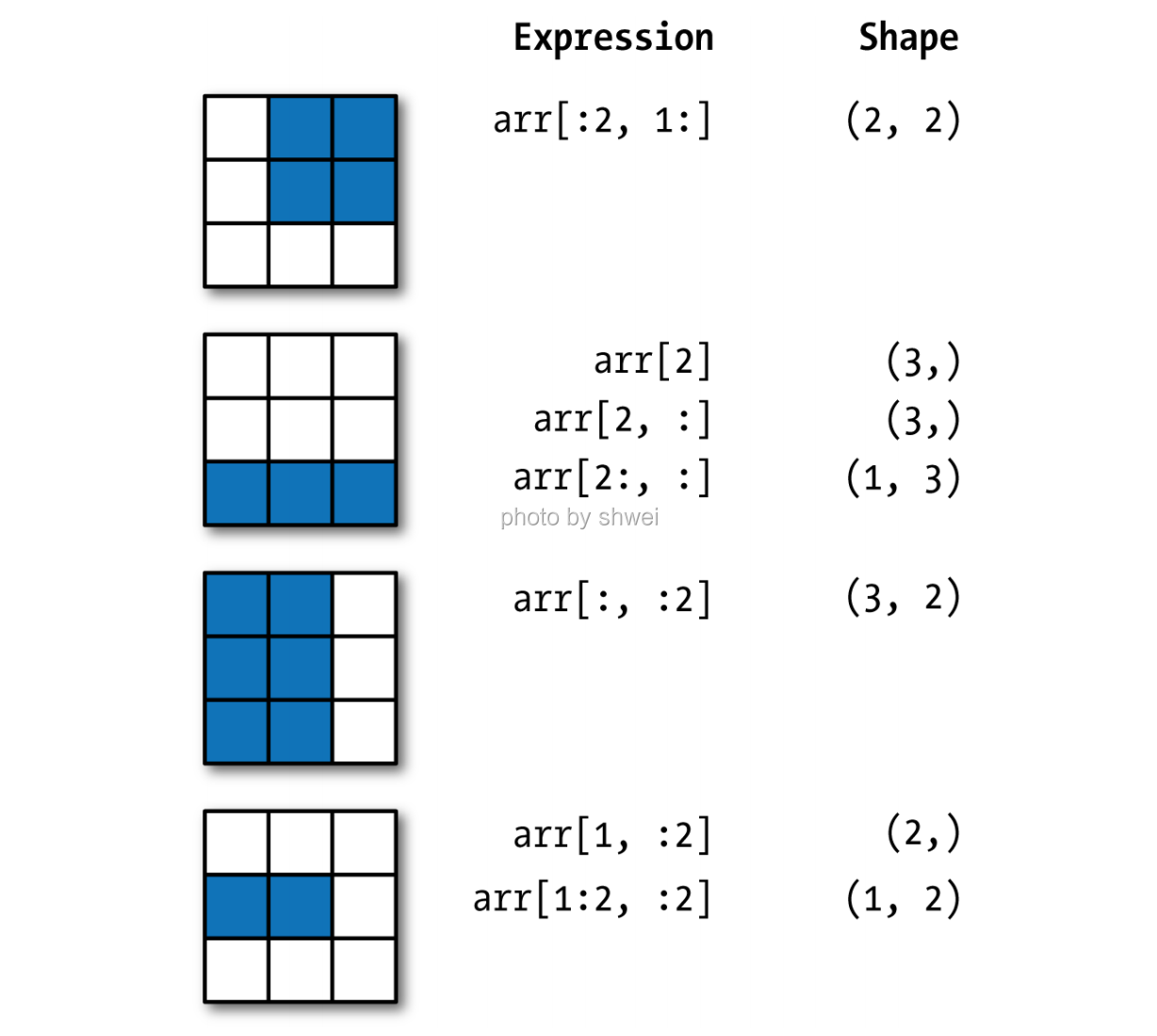
arr2d[:1] # same dimensionality
array([[1, 2, 3]])
arr2d[0] # lower dimensionality
array([1, 2, 3])
arr2d[1,:2]
array([4, 5])
arr2d[:, :1]
array([[1],
[4],
[7]])
arr2d[:2, 1:]
array([[2, 3],
[5, 6]])
arr2d[:2][1:]
array([[4, 5, 6]])
arr_test = arr2d[:2, 1:]
arr_test = 0
arr2d
array([[1, 2, 3],
[4, 5, 6],
[7, 8, 9]])
arr2d[:2, 1:] = 0
arr2d
array([[1, 0, 0],
[4, 0, 0],
[7, 8, 9]])
4.1.5 Boolean Indexing
names = np.array(['Bob', 'Joe', 'Will', 'Bob', 'Will', 'Joe', 'Joe'])
data = np.random.randn(7,4)
names
array(['Bob', 'Joe', 'Will', 'Bob', 'Will', 'Joe', 'Joe'], dtype='<U4')
data
array([[ 2.44492395, -1.52038895, 0.39391998, -0.31685538],
[-0.08518141, 0.38465576, -0.59752249, -1.9550803 ],
[-0.47663211, -0.23931735, 0.71509259, 0.11847852],
[ 1.6580849 , 0.45994775, 0.86858598, -0.7588714 ],
[-0.17831281, -0.3973996 , 0.48140504, -0.20286526],
[ 0.02599393, 0.37176764, -0.83564336, -0.83812596],
[-0.03231511, -1.66515562, 0.04271532, 0.55256624]])
names == 'Bob'
array([ True, False, False, True, False, False, False])
data[names == 'Bob']
array([[ 2.44492395, -1.52038895, 0.39391998, -0.31685538],
[ 1.6580849 , 0.45994775, 0.86858598, -0.7588714 ]])
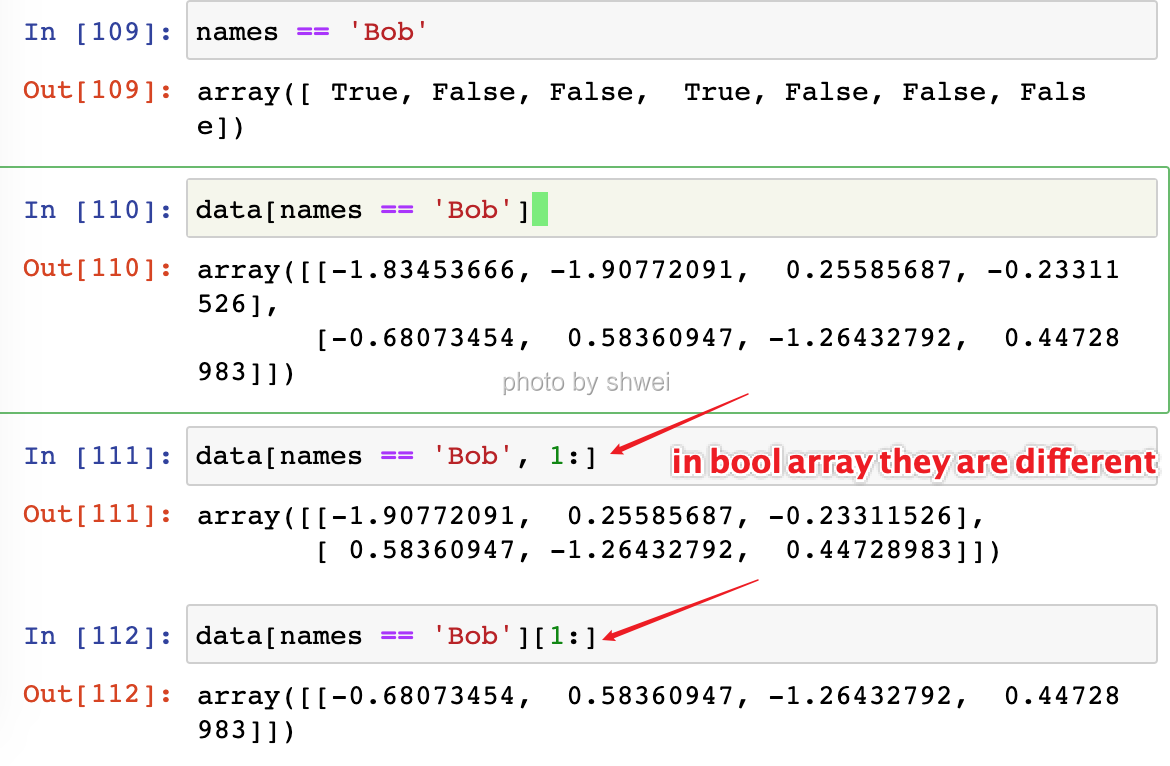
# First dimension fetch data based on boolean
# Second dimension fectch data based on slice
data[names == 'Bob', 1:]
array([[-1.52038895, 0.39391998, -0.31685538],
[ 0.45994775, 0.86858598, -0.7588714 ]])
data[names == 'Bob'][1:]
array([[ 1.6580849 , 0.45994775, 0.86858598, -0.7588714 ]])
data[names == 'Bob', 3]
array([-0.31685538, -0.7588714 ])
To select everything but ‘Bob’, you can either use != or negate the condition using ~
names != 'Bob'
array([False, True, True, False, True, True, True])
data[~(names == 'Bob')]
array([[-0.08518141, 0.38465576, -0.59752249, -1.9550803 ],
[-0.47663211, -0.23931735, 0.71509259, 0.11847852],
[-0.17831281, -0.3973996 , 0.48140504, -0.20286526],
[ 0.02599393, 0.37176764, -0.83564336, -0.83812596],
[-0.03231511, -1.66515562, 0.04271532, 0.55256624]])
Selecting two of the three names to combine mutiple boolean conditions, use boolean arithmetic operators like &(and) and |(or):
Note: The Python keywords and and or do not work with boolean arrays.
mask = (names == 'Bob') | (names == 'Will')
mask
array([ True, False, True, True, True, False, False])
data1 = data[mask] # make a copy
data1
array([[ 2.44492395, -1.52038895, 0.39391998, -0.31685538],
[-0.47663211, -0.23931735, 0.71509259, 0.11847852],
[ 1.6580849 , 0.45994775, 0.86858598, -0.7588714 ],
[-0.17831281, -0.3973996 , 0.48140504, -0.20286526]])
data1[1] = 0
data1
array([[ 2.44492395, -1.52038895, 0.39391998, -0.31685538],
[ 0. , 0. , 0. , 0. ],
[ 1.6580849 , 0.45994775, 0.86858598, -0.7588714 ],
[-0.17831281, -0.3973996 , 0.48140504, -0.20286526]])
data
array([[ 2.44492395, -1.52038895, 0.39391998, -0.31685538],
[-0.08518141, 0.38465576, -0.59752249, -1.9550803 ],
[-0.47663211, -0.23931735, 0.71509259, 0.11847852],
[ 1.6580849 , 0.45994775, 0.86858598, -0.7588714 ],
[-0.17831281, -0.3973996 , 0.48140504, -0.20286526],
[ 0.02599393, 0.37176764, -0.83564336, -0.83812596],
[-0.03231511, -1.66515562, 0.04271532, 0.55256624]])
Note: Selecting data from an array by boolean indexing always creates a copy of the data.
Set all of the negative values in data to 0
data < 0
array([[False, True, False, True],
[ True, False, True, True],
[ True, True, False, False],
[False, False, False, True],
[ True, True, False, True],
[False, False, True, True],
[ True, True, False, False]])
data[data < 0] = 0
data
array([[2.44492395, 0. , 0.39391998, 0. ],
[0. , 0.38465576, 0. , 0. ],
[0. , 0. , 0.71509259, 0.11847852],
[1.6580849 , 0.45994775, 0.86858598, 0. ],
[0. , 0. , 0.48140504, 0. ],
[0.02599393, 0.37176764, 0. , 0. ],
[0. , 0. , 0.04271532, 0.55256624]])
4.1.6 Fancy Indexing
Fancy indexing is a term adpoted by Numpy to describe indexing using integer arrays.
Case 1: Basic Indexing and Slicing
arr[2:]
array([ 2, 3, 4, 12, 100, 12, 8, 9])
arr[0]
0
Case 2: Boolean Indexing
arr = np.array([1,3,4,5])
bool_ = np.array([True, False, True, False], dtype = np.bool)
bool_
array([ True, False, True, False])
arr[bool_]
array([1, 4])
4.1.6.1 Key of Fancy Indexing
Case 3: Fancy Indexing
- Return a copy of original array
- To select out a subset of the rows in a particular order, you can simply pass a list or ndaray of integers specifying the desired order
arr = np.array([1,2,3,4])
arr[[2,3,1,0],] # [2,3,1,0] is a list
array([3, 4, 2, 1])
index_arr = np.array([3,2,0,1])
index_arr
array([3, 2, 0, 1])
arr[index_arr] # index_arr is a ndarray
array([4, 3, 1, 2])
I think Boolean Indexing is a special Fancy Indexing.
arr
array([1, 2, 3, 4])
arr[[1,2]] #(1,) (2,)
array([2, 3])
arr[[-1, -3],] #(-1,) (-3,)
array([4, 2])
arr = np.arange(32).reshape((8,4))
arr
array([[ 0, 1, 2, 3],
[ 4, 5, 6, 7],
[ 8, 9, 10, 11],
[12, 13, 14, 15],
[16, 17, 18, 19],
[20, 21, 22, 23],
[24, 25, 26, 27],
[28, 29, 30, 31]])
# select each dimension
arr[[1,5,7,2],[0,3,1,2]] # (1,0) (5,3) (7,1) (2,2)
array([ 4, 23, 29, 10])
arr[[1,5,7,2]] # (1,:) (5,:) (7,:) (2,:)
array([[ 4, 5, 6, 7],
[20, 21, 22, 23],
[28, 29, 30, 31],
[ 8, 9, 10, 11]])
# Can be understood as: first exchange the second dimension, and then select all the first dimensions
arr[[1,5,7,2]][:, [0,3,1,2]]
array([[ 4, 7, 5, 6],
[20, 23, 21, 22],
[28, 31, 29, 30],
[ 8, 11, 9, 10]])
4.1.7 Transposing Arrays and Swapping Axes
Transposing is a special form of reshaping which similarly returns a view on the underlying data without copying anything.
Transposing: arr[0, 1, 2] = arr.T[2,1,0] After transposing the axes is from dn to d1
arr = np.arange(24).reshape((2,3,4))
arr
array([[[ 0, 1, 2, 3],
[ 4, 5, 6, 7],
[ 8, 9, 10, 11]],
[[12, 13, 14, 15],
[16, 17, 18, 19],
[20, 21, 22, 23]]])
arr[0,1,2]
6
arr size:
arr.T size:
arr_T = arr.T
arr.T # a view of arr
array([[[ 0, 12],
[ 4, 16],
[ 8, 20]],
[[ 1, 13],
[ 5, 17],
[ 9, 21]],
[[ 2, 14],
[ 6, 18],
[10, 22]],
[[ 3, 15],
[ 7, 19],
[11, 23]]])
arr_T[0][1] = 3333
arr
array([[[ 0, 1, 2, 3],
[3333, 5, 6, 7],
[ 8, 9, 10, 11]],
[[ 12, 13, 14, 15],
[3333, 17, 18, 19],
[ 20, 21, 22, 23]]])
arr_T[2][1][0] # arr_T is the array after arr transposing
6
Computing the inner matrix product using np.dot
arr = np.random.randint(1, 4, size=(3,4))
arr
array([[3, 3, 2, 2],
[1, 3, 2, 1],
[2, 2, 3, 1]])
# inner matrix product
np.dot(arr.T, arr)
array([[14, 16, 14, 9],
[16, 22, 18, 11],
[14, 18, 17, 9],
[ 9, 11, 9, 6]])
arr = np.arange(24).reshape((2,3,4))
arr # size 2*3*4
array([[[ 0, 1, 2, 3],
[ 4, 5, 6, 7],
[ 8, 9, 10, 11]],
[[12, 13, 14, 15],
[16, 17, 18, 19],
[20, 21, 22, 23]]])
transpose change axes (0,1,2) to (1,0,2)
So for example the index (1,2,3) will be index(2,1,3)
arr.transpose(1,0,2) # size 2 * 3 * 4
array([[[ 0, 1, 2, 3],
[12, 13, 14, 15]],
[[ 4, 5, 6, 7],
[16, 17, 18, 19]],
[[ 8, 9, 10, 11],
[20, 21, 22, 23]]])
ndarray has the method swapaxes which takes a pair of axis numbers
arr # size 2*3*4
array([[[ 0, 1, 2, 3],
[ 4, 5, 6, 7],
[ 8, 9, 10, 11]],
[[12, 13, 14, 15],
[16, 17, 18, 19],
[20, 21, 22, 23]]])
arr.swapaxes(1,2) # size 2*4*3 return a copy
array([[[ 0, 4, 8],
[ 1, 5, 9],
[ 2, 6, 10],
[ 3, 7, 11]],
[[12, 16, 20],
[13, 17, 21],
[14, 18, 22],
[15, 19, 23]]])
arr[1,2,1]
21
arr.swapaxes(1,2)[1,1,2]
21
4.2 Universal Functions: Fast Element-wise Array Functions
A universal function, or ufunc, is a function that performs elementwise operations on data in ndarrays.You can think of them as fastvectorized wrappers for simple functions.
arr = np.arange(10)
arr
array([0, 1, 2, 3, 4, 5, 6, 7, 8, 9])
np.sqrt(arr)
array([0. , 1. , 1.41421356, 1.73205081, 2. ,
2.23606798, 2.44948974, 2.64575131, 2.82842712, 3. ])
np.exp(arr)
array([1.00000000e+00, 2.71828183e+00, 7.38905610e+00, 2.00855369e+01,
5.45981500e+01, 1.48413159e+02, 4.03428793e+02, 1.09663316e+03,
2.98095799e+03, 8.10308393e+03])
x = np.random.randn(8)
y = np.random.randn(8)
x
array([ 0.60058561, -1.30032727, -1.17669429, -1.16293769, 1.6389972 ,
-0.53815749, -0.14142643, 1.02354324])
y
array([-1.61302358, -0.354913 , -0.1137763 , 0.08010182, 0.07466485,
-1.39663575, 0.77496037, 1.75997402])
np.maximum(x, y)
array([ 0.60058561, -0.354913 , -0.1137763 , 0.08010182, 1.6389972 ,
-0.53815749, 0.77496037, 1.75997402])
arr = np.random.randn(7) * 5
arr
array([ 7.50554328, -0.61609252, 3.00616797, -6.02160661, 3.83440266,
-6.53201864, 7.47775811])
remainder, whole_part = np.modf(arr)
remainder
array([ 0.50554328, -0.61609252, 0.00616797, -0.02160661, 0.83440266,
-0.53201864, 0.47775811])
whole_part
array([ 7., -0., 3., -6., 3., -6., 7.])
Universal Function accept a optional parameter out
arr
array([ 7.50554328, -0.61609252, 3.00616797, -6.02160661, 3.83440266,
-6.53201864, 7.47775811])
np.sqrt(arr, arr)
/usr/local/lib/python3.7/site-packages/ipykernel_launcher.py:1: RuntimeWarning: invalid value encountered in sqrt
"""Entry point for launching an IPython kernel.
array([2.73962466, nan, 1.73383043, nan, 1.95816308,
nan, 2.73454898])
arr
array([2.73962466, nan, 1.73383043, nan, 1.95816308,
nan, 2.73454898])
arr = np.random.randn(10)
arr
array([-0.40366505, -1.98954395, -1.05020216, -0.01818649, -1.08323461,
0.0976977 , -1.45609853, -0.99915274, -1.88666922, -0.10298285])
np.sign(arr)
array([-1., -1., -1., -1., -1., 1., -1., -1., -1., -1.])
arr1 = np.array(range(10))
arr2 = np.arange(1,11)
arr1
array([0, 1, 2, 3, 4, 5, 6, 7, 8, 9])
arr2
array([ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10])
np.subtract(arr1, arr2)
array([-1, -1, -1, -1, -1, -1, -1, -1, -1, -1])
np.copysign(arr, arr2) # copy arr2's sign to arr
array([0.40366505, 1.98954395, 1.05020216, 0.01818649, 1.08323461,
0.0976977 , 1.45609853, 0.99915274, 1.88666922, 0.10298285])
4.2.1 API https://www.geeksforgeeks.org/ , Google, Dash
You don’t need to remember all the function. When you need it, just search the function in Google or Dash.

4.2.2 A listing of available ufuncs
Table 4-3. Unary ufuncs
| Function | Description |
|---|---|
| abs, fabs | Compute the absolute value element-wise for integer, floating point, or complex values.Use fabs as a faster alternative for non-complex-valued data |
| sqrt | Compute the square root of each element. Equivalent to arr ** 0.5 |
| square | Compute the square of each element. Equivalent to arr ** 2 |
| exp | Compute the exponent of each element |
| log, log10, log2, log1p | Natural logarithm (base e), log base 10, log base 2, and log(1 + x), respectively |
| sign | Compute the sign of each element: 1 (positive), 0 (zero), or -1 (negative) |
| ceil | Compute the ceiling of each element, i.e. the smallest integer greater than or equal to each element |
| floor | Compute the floor of each element, i.e. the largest integer less than or equal to each element |
| rint | Round elements to the nearest integer, preserving the dtype |
| modf | Return fractional and integral parts of array as separate array |
| isnan | Return boolean array indicating whether each value is NaN (Not a Number) |
| isfinite, isinf | Return boolean array indicating whether each element is finite (non-inf, non-NaN) or infinite, respectively |
| cos, cosh, sin, sinh, tan, tanh | Regular and hyperbolic trigonometric functions |
| arccos, arccosh, arcsin, arcsinh, arctan, arctanh | Inverse trigonometric functions |
| logical_not | Compute truth value of not x element-wise. Equivalent to -arr |
Table 4-4. Binary universal functions
| Function | Description |
|---|---|
| add | Add corresponding elements in arrays |
| subtract | Subtract elements in second array from first array |
| multiply | Multiply array elements |
| divide, floor_divide | Divide or floor divide (truncating the remainder) |
| power | Raise elements in first array to powers indicated in second array |
| maximum, fmax | Element-wise maximum. fmax ignores NaN |
| minimum, fmin | Element-wise minimum. fmin ignores NaN |
| mod | Element-wise modulus (remainder of division) |
| copysign | Copy sign of values in second argument to values in first argument |
| greater, greater_equal, less, less_equal, equal, not_equal | Perform element-wise comparison, yielding boolean array. Equivalent to infix operators >, >=, <, <=, ==, != |
| logical_and,logical_or, logical_xor | Compute element-wise truth value of logical operation. Equivalent to infix operators &, |, ^ |
4.3 Data Processing Using Arrays
4.3.0 np.meshgrid()
The np.meshgrid function takes two 1D arrays and produces two 2D matrices corresponding to all pairs of (x, y) in the two arrays:
pointx = np.array([1,2,3])
pointy = np.array([-1,-2])
matrices = np.meshgrid(pointx,pointy)
matrices
[array([[1, 2, 3],
[1, 2, 3]]),
array([[-1, -1, -1],
[-2, -2, -2]])]
xs, ys = np.meshgrid(pointx,pointy)
xs
array([[1, 2, 3],
[1, 2, 3]])
ys
array([[-1, -1, -1],
[-2, -2, -2]])
How to understand np.meshgrid()

z = np.sqrt(xs ** 2 + ys ** 2)
z
array([[1.41421356, 2.23606798, 3.16227766],
[2.23606798, 2.82842712, 3.60555128]])
import matplotlib.pyplot as plt
plt.imshow(z, cmap = plt.cm.gray); plt.colorbar()
plt.title("Image plot of $\sqrt{x ^ 2 + y ^ 2}$ for a grid of values")
Text(0.5, 1.0, 'Image plot of $\\sqrt{x ^ 2 + y ^ 2}$ for a grid of values')
[外链图片转存失败,源站可能有防盗链机制,建议将图片保存下来直接上传(img-cQ7da8pB-1583481497049)(output_203_1.png)]
Figure 4-3. Plot of function evaluated on grid
Here I used the matplotlib function imshow to create an image plot
from a 2D array of function values.
4.3.1 Expressing Conditional Logic as Array Operations
The numpy.where function is a vectorized version of the ternary expression x if condtion else y
xarr = np.array([1,2,3,4])
yarr = np.array([-1,-2,-3,-4])
cond = np.array([True, False, True, True])
result = [x if c else y for x, y, c in zip(xarr, yarr, cond)]
result
[1, -2, 3, 4]
This has multiple problems. First, it will not be very fast for large arrays (because all the work is being done in pure Python). Secondly, it will not work with multidimensional arrays.
With np.where you can write this very concisely
result = np.where(cond, xarr, yarr)
result
array([ 1, -2, 3, 4])
The second and third arguments to np.where don’t need to be arrays.A typical use of where in data analysis is to produce a new array of values based on another array.
arr = np.random.randn(4,3)
arr
array([[-0.01139784, 0.40169215, -0.38962953],
[ 0.55216629, 0.48477693, -0.06194546],
[ 0.54855517, 0.17247726, -0.74025193],
[-0.39221758, 0.24833925, 1.15466766]])
np.where(arr > 0, arr, -2)
array([[-2. , 0.40169215, -2. ],
[ 0.55216629, 0.48477693, -2. ],
[ 0.54855517, 0.17247726, -2. ],
[-2. , 0.24833925, 1.15466766]])
The arrays passed to where can be more than just equal sizes array or scales.
4.3.2 Mathematical and Statistical Methods
arr = np.random.randn(5,4) # normally-distributed data
arr
array([[-1.7669711 , 0.23033242, -0.66245059, -1.10571503],
[ 0.17697343, 0.09412475, 2.31777073, -0.44555725],
[-0.62426573, 1.75648072, 0.95977683, -0.61619976],
[-0.49642721, 0.45912091, 1.26163226, 0.06395789],
[ 1.66048345, 0.34056885, -1.54478641, -1.04533225]])
arr = np.arange(15).reshape(5, 3)
arr
array([[ 0, 1, 2],
[ 3, 4, 5],
[ 6, 7, 8],
[ 9, 10, 11],
[12, 13, 14]])
arr.mean() # Average of all numbers
7.0
arr.sum() # Sum of all numbers
105
Functions like mean and sum take an optional axis argument
which computes the statistic over the given axis, resulting in an array
with one fewer dimension, or compressing the given axis
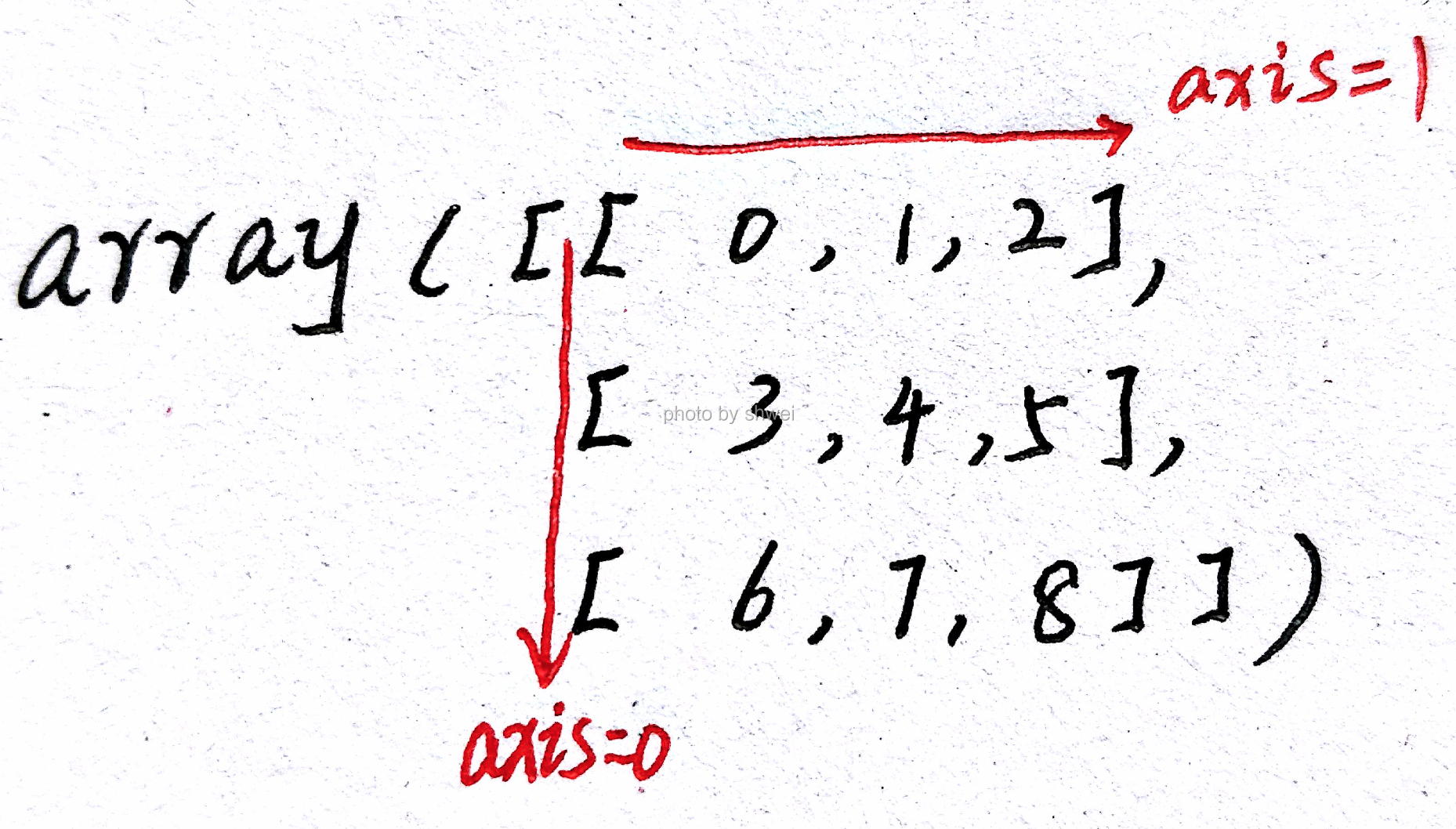
arr
array([[ 0, 1, 2],
[ 3, 4, 5],
[ 6, 7, 8],
[ 9, 10, 11],
[12, 13, 14]])
# arr[3][1] == 10
# axis 1 is the direction 0->1->2 here
arr.mean(axis = 1) # compress axis 1 and here is (5, 3)‘s 3
array([ 1., 4., 7., 10., 13.])
arr.sum(axis = 0)
array([30, 35, 40])
arr.sum(0)
array([30, 35, 40])
arr.min(1) # dimension reduced
array([ 0, 3, 6, 9, 12])
arr.argmax(1) # Indices of maximum elements, respective
array([2, 2, 2, 2, 2])
arr = np.arange(10)
arr
array([0, 1, 2, 3, 4, 5, 6, 7, 8, 9])
arr.cumsum()
array([ 0, 1, 3, 6, 10, 15, 21, 28, 36, 45])
arr = np.arange(9).reshape(3,3)
arr
array([[0, 1, 2],
[3, 4, 5],
[6, 7, 8]])
arr.cumsum(0)
array([[ 0, 1, 2],
[ 3, 5, 7],
[ 9, 12, 15]])
arr.cumprod(axis = 1)
array([[ 0, 0, 0],
[ 3, 12, 60],
[ 6, 42, 336]])
Table 4-5. Basic array statistical methods
| Method | Description |
|---|---|
| sum | Sum of all the elements in the array or along an axis. Zero-length arrays have sum 0. |
| mean | Arithmetic mean. Zero-length arrays have NaN mean. |
| std, var | Standard deviation and variance, respectively, with optional degrees of freedom adjustment (default denominator n). |
| min, max | Minimum and maximum. |
| argmin, argmax | Indices of minimum and maximum elements, respectively. |
| cumsum | Cumulative sum of elements starting from 0 |
| cumprod | Cumulative product of elements starting from 1 |
4.3.3 Methods for Boolean Arrays
Boolean values are coerced to 1(True) and 0(False) in the above methods.
arr = np.random.randn(100)
(arr > 0).sum() # Number of positive values
46
There are two additional methods, any and all, useful especially for boolean
arrays.
any tests whether one or more values in an array is True, while all checks if every value
is True
bools = np.array([True, False, False, False])
bools.any()
True
bools.all()
False
bools.sum()
1
4.3.4 Sorting
Like Python’s built-in list type, NumPy arrays can be sorted in-place using the sort method
arr = np.random.randn(8)
arr
array([-0.05645386, -0.36224588, -1.29498701, -0.30476409, -0.46770012,
1.20385542, 1.47953717, -0.20565266])
arr.sort()
arr
array([-1.29498701, -0.46770012, -0.36224588, -0.30476409, -0.20565266,
-0.05645386, 1.20385542, 1.47953717])
4.3.5 Unique and Other Set Logic
NumPy has some basic set operations for one-dimensional ndarrays. Probably
the most commonly used one is np.unique, which returns the sorted unique values in an array
names = np.array(['Bob', 'Will','Joe', 'Bob', 'Will'])
np.unique(names)
array(['Bob', 'Joe', 'Will'], dtype='<U4')
names
array(['Bob', 'Will', 'Joe', 'Bob', 'Will'], dtype='<U4')
ints = np.array([3,3,3,2,2,1,2,3,5])
np.unique(ints)
array([1, 2, 3, 5])
Constract np.unique with the pure Python alternative
sorted(set(names))
['Bob', 'Joe', 'Will']
Another function, np.in1d, tests membership of the values in one array in another, returning a boolean array.
values = np.array([3,2,31,44,34,5])
np.in1d(values, [2,3])
array([ True, True, False, False, False, False])
Table 4-6. Array set operations
| Method | Description |
|---|---|
| unique(x) | Compute the sorted, unique elements in x |
| intersect1d(x, y) | Compute the sorted, common elements in x and y |
| union1d(x, y) | Compute the sorted union of elements |
| in1d(x, y) | Compute a boolean array indicating whether each element of x is contained in y |
| setdiff1d(x, y) | Set difference, elements in x that are not in y |
| setxor1d(x, y) | Set symmetric differences; elements that are in either of the arrays, but not both |
4.4 File Input and Output with Arrays
NumPy is able to save and load data to and from disk either in text or binary format.
In later chapters you will learn about tools in pandas for reading tabular data into
memory.
4.5 Linear Algebra
Linear algebra, like matrix multiplication, decompostions, determinants, and other square matrix math, is
an important part of any array library.
Unlike some languages like MATLAB, multiplying two-dimensional arrays with * is an element-wise product instead of a matrix dot product.
As such, there is a function dot, both an array
method, and a function in the numpy namespace
x = np.array([[1,2,3],[4,5,6]])
x
array([[1, 2, 3],
[4, 5, 6]])
x * x
array([[ 1, 4, 9],
[16, 25, 36]])
y = np.arange(6).reshape(3,2)
y
array([[0, 1],
[2, 3],
[4, 5]])
x.dot(y) # ndarray function
array([[16, 22],
[34, 49]])
np.dot(x,y) # a function in numpy namespace
array([[16, 22],
[34, 49]])
x
array([[1, 2, 3],
[4, 5, 6]])
x @ np.ones(3) # @ is also used in a matrix dot product
array([ 6., 15.])
numpy.linalg(linear algebra) has a standard set of matrix decompositions and things like inverse and determinant. These are implemented under the hood using the same industry-standard Fortran libraries used in other languages like MATLAB and R, such as like BLAS,LAPACK, or possibly(depending on your NumPy build) the Intel MKL
更新标准写法是:from numpy.linalg import inv, qr,这样求逆就直接写inv(matrix)
import numpy.linalg as npla # npla是我自己取的名,不规范
matrix = np.random.randn(4,4)
npla.inv(matrix)
array([[-46.76938498, -86.35815947, 129.96228838, -32.95425663],
[-22.31060821, -41.56709931, 62.01812792, -15.25370697],
[-20.35569284, -37.11177381, 54.70106286, -14.5358842 ],
[-30.82139292, -54.49869152, 81.79616404, -21.54068957]])
arr = matrix.dot(npla.inv(matrix)) # np.dot(matrix, npla.inv(matrix))
arr
array([[ 1.00000000e+00, -5.73321716e-15, -4.64130813e-15,
-1.34965101e-16],
[ 1.03102760e-15, 1.00000000e+00, -1.13084465e-14,
6.16824551e-16],
[-6.92801263e-15, -9.06342203e-15, 1.00000000e+00,
-3.46276746e-15],
[ 1.61273987e-17, -4.13259316e-15, -9.54630608e-15,
1.00000000e+00]])
np.round(arr, 2) # decimals=2
array([[ 1., -0., -0., -0.],
[ 0., 1., -0., 0.],
[-0., -0., 1., -0.],
[ 0., -0., -0., 1.]])
Table 4-7. Commonly-used numpy.linalg functions
| Function | Description |
|---|---|
| diag | Return the diagonal (or off-diagonal) elements of a square matrix as a 1D array, or convert a 1D array into a square matrix with zeros on the off-diagonal |
| dot | Matrix multiplication |
| trace | Compute the sum of the diagonal elements |
| det | Compute the matrix determinant |
| eig | Compute the eigenvalues and eigenvectors of a square matrix |
| inv | Compute the inverse of a square matrix |
| pinv | Compute the Moore-Penrose pseudo-inverse inverse of a square matrix |
| qr | Compute the QR decomposition |
| svd | Compute the singular value decomposition (SVD) |
| solve | Solve the linear system Ax = b for x, where A is a square matrix |
| lstsq | Compute the least-squares solution to y = Xb |
4.6 Random Number Generation
The numpy.random module supplements the built-in Python random with functions for efficiently generating whole arrays of sample values from many kinds of probabiliry distributions.
Table 4-8. Partial list of numpy.random functions
| Function | Description |
|---|---|
| seed | Seed the random number generator |
| permutation | Return a random permutation of a sequence, or return a permuted range |
| shuffle | Randomly permute a sequence in place |
| rand | Draw samples from a uniform distribution |
| randint | Draw random integers from a given low-to-high range |
| randn | Draw samples from a normal distribution with mean 0 and standard deviation 1 (MATLAB-like interface) |
| binomial | Draw samples a binomial distribution |
| normal | Draw samples from a normal (Gaussian) distribution |
| beta | Draw samples from a beta distribution |
| chisquare | Draw samples from a chi-square distribution |
| gamma | Draw samples from a gamma distribution |
| uniform | Draw samples from a uniform [0, 1) distribution |
samples = np.random.normal(size = (4,4))
np.round(samples,4)
array([[-0.4422, -0.1181, -1.1577, 0.4524],
[ 0.923 , 1.8407, 0.1605, -0.9159],
[ 0.7133, -0.0398, -1.3522, -1.1874],
[-0.4133, 0.7873, -0.9728, -0.9143]])
Python’s built-in random module by constrast only samples one value at a time. As you can see from this benchmark, numpy.random is well over an order of magnitude faster for generating very large samples
from random import normalvariate
N = 1000000
%timeit sample = [normalvariate(0,1) for _ in range(N)]
1.16 s ± 99.4 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
%timeit np.random.normal(size = N)
33 ms ± 1.39 ms per loop (mean ± std. dev. of 7 runs, 10 loops each)
They’re actually what’s known as " pseudo random numbers, " generated by algorithms.

4.6.1 random.random((m,n))
The function parameter is a tuple
Generate random numbers from 0 to 1
import numpy as np
data = np.random.random((2,5))
# Show the variable
data
array([[0.87481527, 0.11475954, 0.52484471, 0.86935258, 0.09528812],
[0.15106181, 0.12227048, 0.81898961, 0.33022438, 0.94330205]])
# Print the variable
print(data)
[[0.87481527 0.11475954 0.52484471 0.86935258 0.09528812]
[0.15106181 0.12227048 0.81898961 0.33022438 0.94330205]]
data * 10
array([[8.74815275, 1.14759543, 5.24844707, 8.69352576, 0.9528812 ],
[1.51061807, 1.22270479, 8.18989614, 3.30224382, 9.43302047]])
print(data + data)
[[1.74963055 0.22951909 1.04968941 1.73870515 0.19057624]
[0.30212361 0.24454096 1.63797923 0.66044876 1.88660409]]
ndim: the first dimension
data.ndim
2
shape: a tuple indicating the size of each dimension
data.shape
(2, 5)
data_ = np.array([1,2,3])
data_.shape # (3,) Only have one dimension
(3,)
dtype: decribe the data type of the array
data.dtype
dtype('float64')
data2 = np.random.random(9)
data2
array([0.0155825 , 0.80244718, 0.14901417, 0.42191698, 0.60644615,
0.37694115, 0.2821992 , 0.97835977, 0.08436808])
4.6.2 random.randint(1,8, size = (3,2))
Generate random integer from 1 to 8, left close and right open
data_randint = np.random.randint(1,8,size = (3, 2))
data_randint
array([[7, 6],
[7, 7],
[1, 2]])
4.6.3 random.randn(d1, d2, d3, …)
randn generates a matrix filled with random floats sampled from a univarite “normal” distribution of mean 0 and variance 1
Notes:
For random samples from
,use:
sigma * np.random.randn(d1, ) + mu
# N(1, 6.25)
data_normal_distribution = 1 * np.random.randn(2,3) + 2.5
data_normal_distribution
array([[4.03463856, 5.86698859, 2.59342223],
[2.18876421, 1.66281754, 1.51047393]])
NumPy Summay
The biggest benefit of NumPy arrays is the use of simple array expressions to complete a variety of data manipulation tasks without the need to write some loops.
- Basic Indexing, Slicing, arr.T return view
- Boolean Indexing, Fancy Indexing, arr.swapaxes return copy
arr2d
array([[1, 0, 0],
[4, 0, 0],
[7, 8, 9]])
arr_test = arr2d[:2, 1:] # return view
arr_test = 0 # This will change the original arr2d
arr2d
array([[1, 0, 0],
[4, 0, 0],
[7, 8, 9]])
,locate the dimension:keep in same dimension (Slicing)[2,3,1]choose some in this dimension (Fancy Indexing)
arr2d[1,0]
4
arr2d[1,]
array([4, 0, 0])
arr2d[:2,0]
array([1, 4])
arr2d[:2,:1]
array([[1],
[4]])
arr2d[[2,1]]
array([[7, 8, 9],
[4, 0, 0]])
How to locate number 5
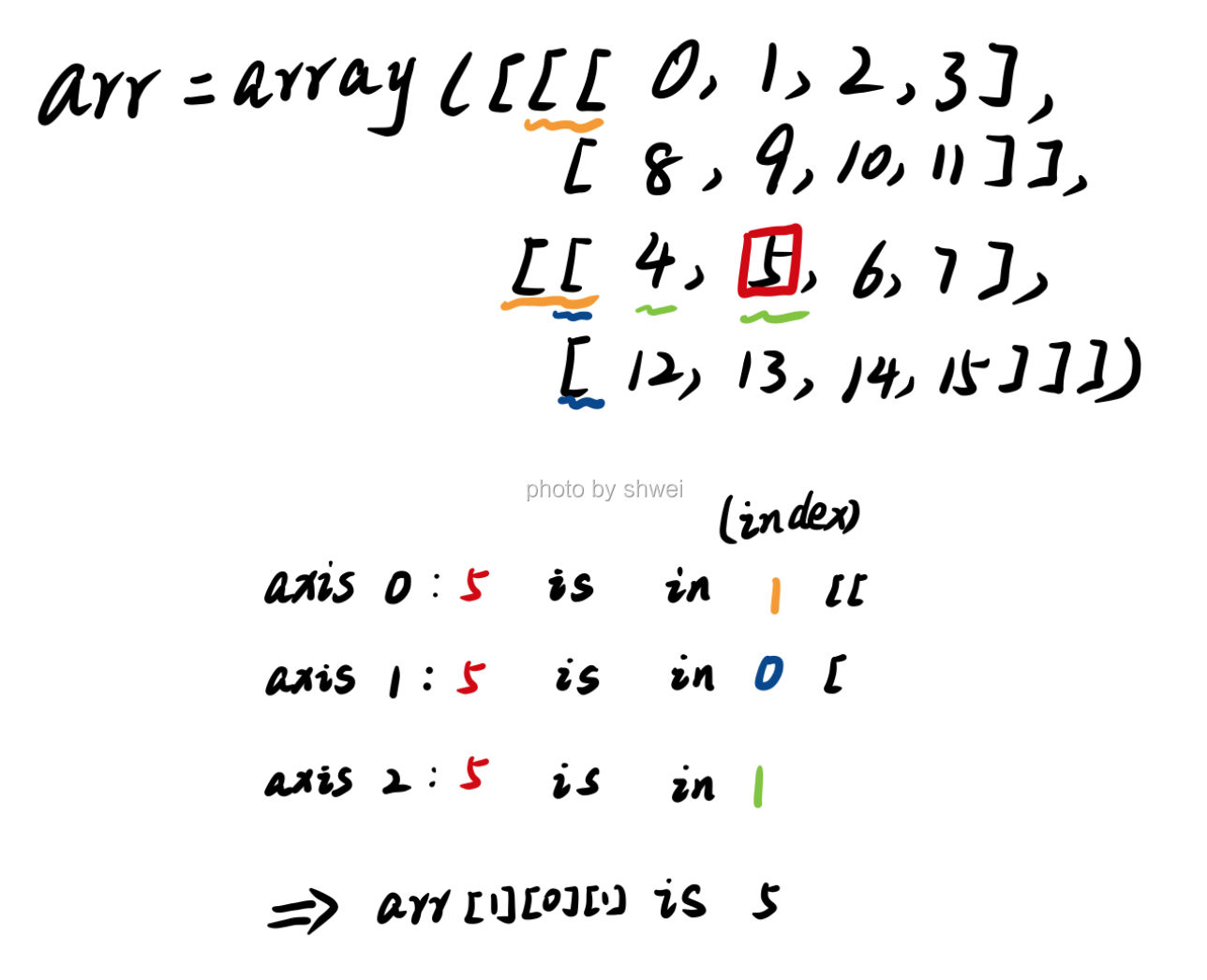
Change axis
Example 1:
arr size: 2∗3∗4
arr.T size: 4∗3∗2
old:(0,1,2) = 6
now:(2,1,0) = 6