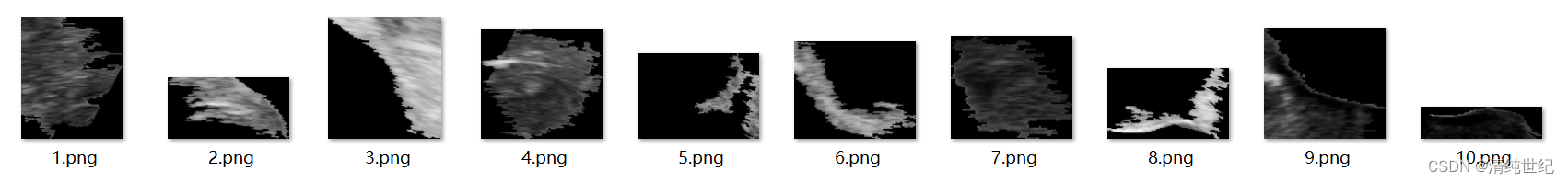
SLIC スーパーピクセル セグメンテーションとセグメント化されたスーパーピクセル ブロックの保存:
# https://github.com/LarkMi/SLIC/blob/main/SLIC.py
import skimage
from skimage.segmentation import slic,mark_boundaries
from skimage import io
import matplotlib.pyplot as plt
from PIL import Image, ImageEnhance
import numpy as np
import cv2
#
# np.set_printoptions(threshold=np.inf)
path = 'C:\\Users\\Administrator\\Desktop\\SLIC\\'
img_name = 'test.png'
img = io.imread(path + img_name,as_gray=True) #as_gray是灰度读取,得到的是归一化值
segments = slic(img, n_segments=10, compactness=0.2,start_label = 1)#进行SLIC分割
out=mark_boundaries(img,segments)
out = out*255 #io的灰度读取是归一化值,若读取彩色图片去掉该行
img3 = Image.fromarray(np.uint8(out))
img3.show()
seg_img_name = 'seg.png'
img3.save(path +'\\' +seg_img_name)#显示并保存加上分割线后的图片
maxn = max(segments.reshape(int(segments.shape[0]*segments.shape[1]),))
for i in range(1,maxn+1):
a = np.array(segments == i)
b = img * a
w,h = [],[]
for x in range(b.shape[0]):
for y in range(b.shape[1]):
if b[x][y] != 0:
w.append(x)
h.append(y)
c = b[min(w):max(w),min(h):max(h)]
c = c*255
d = c.reshape(c.shape[0],c.shape[1],1)
e = np.concatenate((d,d),axis=2)
e = np.concatenate((e,d),axis=2)
img2 = Image.fromarray(np.uint8(e))
img2.save(path +'\\'+str(i)+'.png')
print('已保存第' + str(i) + '张图片')
wid,hig = [],[]
img = io.imread(path+'\\'+seg_img_name)
for i in range(1,maxn+1):
w,h = [],[]
for x in range(segments.shape[0]):
for y in range(segments.shape[1]):
if segments[x][y] == i:
w.append(x)
h.append(y)
font=cv2.FONT_HERSHEY_SIMPLEX#使用默认字体
#print((min(w),min(h)))
img=cv2.putText(img,str(i),(h[int(len(h)/(2))],w[int(len(w)/2)]),font,1,(255,255,255),2)#添加文字,1.2表示字体大小,(0,40)是初始的位置,(255,255,255)表示颜色,2表示粗细
img = Image.fromarray(np.uint8(img))
img.show()
img.save(path +'\\'+seg_img_name+'_label.png')
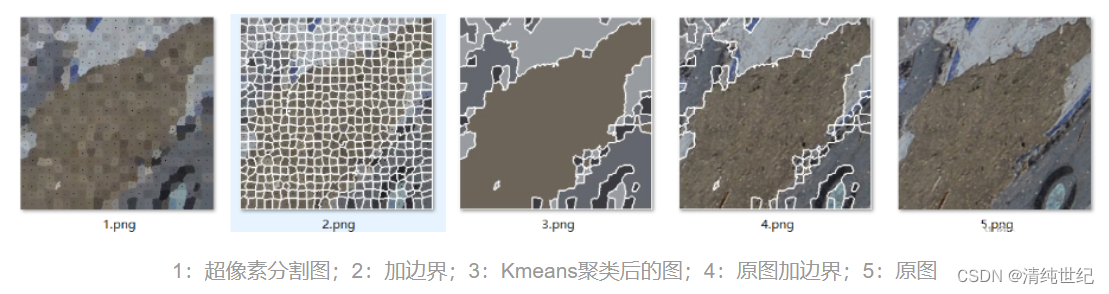
自作のスーパーピクセル セグメンテーション コード:
- k-means アルゴリズムの結果のラベル付けを容易にするために、label 属性を Cluster クラスに追加しました。
- SLICProcessorクラスのsave_current_imageメソッドに、3||4.pngのような画像を生成できる枠線増加部分を追加しました。
- 新しいクラス メソッドgenerate_result() を追加しました。ユーザー パラメータ K は、設定された Kmeans アルゴリズムのクラスタ数であり、クラスタ数に応じてマージする領域を選択します。
- 元のコードでは、画像チャンネル数が異なるため、jpg と png 画像を同時に読み込むことができないため、小さな変更で対応できます。
# https://gitee.com/xu-qiyu/MyProject/tree/master/opencv/超像素分割
from copy import copy, deepcopy
import math
from cv2 import CHAIN_APPROX_NONE, RETR_LIST, imshow, merge, findContours, waitKey
from skimage import io, color
import numpy as np
from tqdm import trange
from tqdm import tqdm
from sklearn.cluster import KMeans
class Cluster(object):
cluster_index = 1
def __init__(self, h, w, l=0, a=0, b=0):
self.update(h, w, l, a, b)
self.pixels = []
self.no = self.cluster_index
self.label = 0
Cluster.cluster_index += 1
def update(self, h, w, l, a, b):
self.h = h
self.w = w
self.l = l
self.a = a
self.b = b
def __str__(self):
return "{},{}:{} {} {} ".format(self.h, self.w, self.l, self.a, self.b)
def __repr__(self):
return self.__str__()
class SLICProcessor(object):
@staticmethod
def open_image(path):
"""
Return:
3D array, row col [LAB]
"""
rgb = io.imread(path)
if path[-3:] == 'png':
lab_arr = color.rgb2lab(rgb[:, :, 0:3])
else:
lab_arr = color.rgb2lab(rgb)
return lab_arr
@staticmethod
def save_lab_image(path, lab_arr):
"""
Convert the array to RBG, then save the image
:param path:
:param lab_arr:
:return:
"""
rgb_arr = color.lab2rgb(lab_arr)
io.imsave(path, rgb_arr)
def make_cluster(self, h, w):
h = int(h)
w = int(w)
return Cluster(h, w,
self.data[h][w][0],
self.data[h][w][1],
self.data[h][w][2])
def __init__(self, filename, K, M):
self.K = K
self.M = M
self.data = self.open_image(filename)
self.image_height = self.data.shape[0]
self.image_width = self.data.shape[1]
self.N = self.image_height * self.image_width
self.S = int(math.sqrt(self.N / self.K))
self.clusters = []
self.label = {}
self.dis = np.full((self.image_height, self.image_width), np.inf)
def init_clusters(self):
h = self.S // 2
w = self.S // 2
while h < self.image_height:
while w < self.image_width:
self.clusters.append(self.make_cluster(h, w))
w += self.S
w = self.S // 2
h += self.S
def get_gradient(self, h, w):
if w + 1 >= self.image_width:
w = self.image_width - 2
if h + 1 >= self.image_height:
h = self.image_height - 2
gradient = self.data[h + 1][w + 1][0] - self.data[h][w][0] + \
self.data[h + 1][w + 1][1] - self.data[h][w][1] + \
self.data[h + 1][w + 1][2] - self.data[h][w][2]
return gradient
def move_clusters(self):
for cluster in self.clusters:
cluster_gradient = self.get_gradient(cluster.h, cluster.w)
for dh in range(-1, 2):
for dw in range(-1, 2):
_h = cluster.h + dh
_w = cluster.w + dw
new_gradient = self.get_gradient(_h, _w)
if new_gradient < cluster_gradient:
cluster.update(_h, _w, self.data[_h][_w][0], self.data[_h][_w][1], self.data[_h][_w][2])
cluster_gradient = new_gradient
def assignment(self):
for cluster in tqdm(self.clusters):
for h in range(cluster.h - 2 * self.S, cluster.h + 2 * self.S):
if h < 0 or h >= self.image_height: continue
for w in range(cluster.w - 2 * self.S, cluster.w + 2 * self.S):
if w < 0 or w >= self.image_width: continue
L, A, B = self.data[h][w]
Dc = math.sqrt(
math.pow(L - cluster.l, 2) +
math.pow(A - cluster.a, 2) +
math.pow(B - cluster.b, 2))
Ds = math.sqrt(
math.pow(h - cluster.h, 2) +
math.pow(w - cluster.w, 2))
D = math.sqrt(math.pow(Dc / self.M, 2) + math.pow(Ds / self.S, 2))
if D < self.dis[h][w]:
if (h, w) not in self.label:
self.label[(h, w)] = cluster
cluster.pixels.append((h, w))
else:
self.label[(h, w)].pixels.remove((h, w))
self.label[(h, w)] = cluster
cluster.pixels.append((h, w))
self.dis[h][w] = D
a = 1
def update_cluster(self):
for cluster in self.clusters:
sum_h = sum_w = number = 0
for p in cluster.pixels:
sum_h += p[0]
sum_w += p[1]
number += 1
_h = int(sum_h / number)
_w = int(sum_w / number)
cluster.update(_h, _w, self.data[_h][_w][0], self.data[_h][_w][1], self.data[_h][_w][2])
def save_current_image(self, name):
image_arr = np.copy(self.data)
# imshow("r", image_arr)
for cluster in self.clusters:
for p in cluster.pixels:
image_arr[p[0]][p[1]][0] = cluster.l
image_arr[p[0]][p[1]][1] = cluster.a
image_arr[p[0]][p[1]][2] = cluster.b
image_arr[cluster.h][cluster.w][0] = 0
image_arr[cluster.h][cluster.w][1] = 0
image_arr[cluster.h][cluster.w][2] = 0
self.save_lab_image("1.png", image_arr)
mask_r = np.ones((self.image_height, self.image_width), np.uint8)*0
for cluster in self.clusters:
mask = np.ones((self.image_height, self.image_width), np.uint8)*0
for x in cluster.pixels:
mask[x[0], x[1]] = 255
contours, _ = findContours(mask, RETR_LIST, CHAIN_APPROX_NONE)
for contour in contours:
for i in contour:
mask_r[i[0][1], i[0][0]] = 255
for x in range(self.image_height):
for y in range(self.image_width):
if mask_r[x, y] == 255:
image_arr[x, y] = [100, 0, 0]
self.save_lab_image("2.png", image_arr)
def iterate_10times(self):
self.init_clusters()
self.move_clusters()
# for i in trange(10):
self.assignment()
self.update_cluster()
# name = 'lenna_M{m}_K{k}_loop{loop}.png'.format(loop=0, m=self.M, k=self.K)
self.save_current_image("123")
def generate_result(self, K):
clusters = deepcopy(self.clusters)
temp_img = [[x.l, x.a, x.b] for x in clusters]
kmeans = KMeans(n_clusters=K,random_state=3).fit(temp_img)
for i in range(len(self.clusters)):
self.clusters[i].label = kmeans.labels_[i]
mask = np.ones((self.image_height, self.image_width))
img = merge([mask, mask, mask])
for cluster in self.clusters:
for pixel in cluster.pixels:
img[pixel[0], pixel[1]] = kmeans.cluster_centers_[cluster.label]
for num in range(K):
mask = np.ones((self.image_height, self.image_width), np.uint8)*0
for cluster in self.clusters:
if cluster.label == num:
for pixel in cluster.pixels:
mask[pixel[0], pixel[1]] = 255
contours, _ = findContours(mask, RETR_LIST, CHAIN_APPROX_NONE)
for contour in contours:
# if len(contour)<200: continue#可以注释掉这行获取小区域的分割
for i in contour:
img[i[0][1], i[0][0]] = [100, 0, 0]
self.data[i[0][1], i[0][0]] = [100, 0, 0]
self.save_lab_image("3.png", img)
self.save_lab_image("4.png", self.data)
if __name__ == '__main__':
p = SLICProcessor('5.png', 500, 40)
p.iterate_10times()
p.generate_result(4)
waitKey()
Pythonのslic関数:
def slic(image, n_segments=100, compactness=10., max_iter=10, sigma=0,
spacing=None, multichannel=True, convert2lab=None,
enforce_connectivity=True, min_size_factor=0.5, max_size_factor=3,
slic_zero=False):
"""Segments image using k-means clustering in Color-(x,y,z) space.
Parameters
----------
image : 2D, 3D or 4D ndarray
Input image, which can be 2D or 3D, and grayscale or multichannel
(see `multichannel` parameter).
n_segments : int, optional
The (approximate) number of labels in the segmented output image.
compactness : float, optional
控制颜色和空间之间的平衡,约高越方块,和图关系密切,最好先确定指数级别,再微调
Balances color proximity and space proximity. Higher values give
more weight to space proximity, making superpixel shapes more
square/cubic. In SLICO mode, this is the initial compactness.
This parameter depends strongly on image contrast and on the
shapes of objects in the image. We recommend exploring possible
values on a log scale, e.g., 0.01, 0.1, 1, 10, 100, before
refining around a chosen value.
max_iter : int, optional
最大k均值迭代次数
Maximum number of iterations of k-means.
sigma : float or (3,) array-like of floats, optional
图像每个维度进行预处理时的高斯平滑核宽。若给定为标量值,则同一个值运用到各个维度。0意味
着不平滑。如果“sigma”是标量的,并且提供了手动体素间距,则自动缩放它(参见注释部分)。
Width of Gaussian smoothing kernel for pre-processing for each
dimension of the image. The same sigma is applied to each dimension in
case of a scalar value. Zero means no smoothing.
Note, that `sigma` is automatically scaled if it is scalar and a
manual voxel spacing is provided (see Notes section).
spacing : (3,) array-like of floats, optional
代表沿着图像每个维度的体素空间。默认情况下,slic假定均匀的空间(沿x,y,z轴相同的体素分辨
率),这个参数控制在k均值聚类中各轴距离的权重
The voxel spacing along each image dimension. By default, `slic`
assumes uniform spacing (same voxel resolution along z, y and x).
This parameter controls the weights of the distances along z, y,
and x during k-means clustering.
multichannel : bool, optional
二进制参数,代表图像的最后一个轴代表多通道还是另一个空间维度
Whether the last axis of the image is to be interpreted as multiple
channels or another spatial dimension.
convert2lab : bool, optional
二进制参数,判断输入需要在分割之前转到LAB颜色空间。输入必须是RGB。当多通道参数为True,
输入图片的通道数为3时,该参数默认为True
Whether the input should be converted to Lab colorspace prior to
segmentation. The input image *must* be RGB. Highly recommended.
This option defaults to ``True`` when ``multichannel=True`` *and*
``image.shape[-1] == 3``.
enforce_connectivity: bool, optional
二进制参数,控制生成的分割块连接或不连接
Whether the generated segments are connected or not
min_size_factor: float, optional
与分割目标数有关的要删去的最小分割块比率,(大概是小于长*宽*高/目标数量 的分割结果会被融
合掉)
Proportion of the minimum segment size to be removed with respect
to the supposed segment size ```depth*width*height/n_segments```
max_size_factor: float, optional
最大融合比率上限
Proportion of the maximum connected segment size. A value of 3 works
in most of the cases.
slic_zero: bool, optional
不知所谓的零参数
Run SLIC-zero, the zero-parameter mode of SLIC. [2]_
Returns
-------
labels : 2D or 3D array
Integer mask indicating segment labels.
Raises
------
ValueError
If ``convert2lab`` is set to ``True`` but the last array
dimension is not of length 3.
Notes
-----
* If `sigma > 0`, the image is smoothed using a Gaussian kernel prior to
segmentation.
* If `sigma` is scalar and `spacing` is provided, the kernel width is
divided along each dimension by the spacing. For example, if ``sigma=1``
and ``spacing=[5, 1, 1]``, the effective `sigma` is ``[0.2, 1, 1]``. This
ensures sensible smoothing for anisotropic images.
如果有平滑参数sigma和体素空间参数spacing,那么空间体素参数会对平滑参数有平分的影响,比如
1/[5,1,1]=[0.2,1,1]
* The image is rescaled to be in [0, 1] prior to processing.
图像在预处理之前会被处理为[0,1]之间的标量
* Images of shape (M, N, 3) are interpreted as 2D RGB images by default. To
interpret them as 3D with the last dimension having length 3, use
`multichannel=False`.
(M,N,3)的图像默认为2维(RGB的图像),要想被理解为3维图需要设置多通道参数=False
References
----------
.. [1] Radhakrishna Achanta, Appu Shaji, Kevin Smith, Aurelien Lucchi,
Pascal Fua, and Sabine Süsstrunk, SLIC Superpixels Compared to
State-of-the-art Superpixel Methods, TPAMI, May 2012.
.. [2] http://ivrg.epfl.ch/research/superpixels#SLICO
Examples
--------
>>> from skimage.segmentation import slic
>>> from skimage.data import astronaut
>>> img = astronaut()
>>> segments = slic(img, n_segments=100, compactness=10)
Increasing the compactness parameter yields more square regions:
>>> segments = slic(img, n_segments=100, compactness=20)
"""
###############################################干正事啦
image = img_as_float(image)
is_2d = False
#2D灰度图
if image.ndim == 2:
# 2D grayscale image
image = image[np.newaxis, ..., np.newaxis]
is_2d = True
#比如2D RGB的图
elif image.ndim == 3 and multichannel:
# Make 2D multichannel image 3D with depth = 1
image = image[np.newaxis, ...]
is_2d = True
#比如3D图
elif image.ndim == 3 and not multichannel:
# Add channel as single last dimension
image = image[..., np.newaxis]
#控制聚类时各轴权重
if spacing is None:
spacing = np.ones(3)
elif isinstance(spacing, (list, tuple)):
spacing = np.array(spacing, dtype=np.double)
#高斯平滑
if not isinstance(sigma, coll.Iterable):
sigma = np.array([sigma, sigma, sigma], dtype=np.double)
sigma /= spacing.astype(np.double)#有可能发生的体素除
elif isinstance(sigma, (list, tuple)):
sigma = np.array(sigma, dtype=np.double)
#高斯滤波处
if (sigma > 0).any():
# add zero smoothing for multichannel dimension
sigma = list(sigma) + [0]
image = ndi.gaussian_filter(image, sigma)
#多通道RGB图且需要转lab,用rab2lab即可实现
if multichannel and (convert2lab or convert2lab is None):
if image.shape[-1] != 3 and convert2lab:
raise ValueError("Lab colorspace conversion requires a RGB image.")
elif image.shape[-1] == 3:
image = rgb2lab(image)
depth, height, width = image.shape[:3]
# initialize cluster centroids for desired number of segments
#为实现目标分割块数,初始化聚类中心。
#grid_* 相当于index
#slices是根据目标数量分的块,有取整需要
grid_z, grid_y, grid_x = np.mgrid[:depth, :height, :width]
slices = regular_grid(image.shape[:3], n_segments)
step_z, step_y, step_x = [int(s.step if s.step is not None else 1)
for s in slices]
segments_z = grid_z[slices]
segments_y = grid_y[slices]
segments_x = grid_x[slices]
segments_color = np.zeros(segments_z.shape + (image.shape[3],))
segments = np.concatenate([segments_z[..., np.newaxis],
segments_y[..., np.newaxis],
segments_x[..., np.newaxis],
segments_color],
axis=-1).reshape(-1, 3 + image.shape[3])
segments = np.ascontiguousarray(segments)
# we do the scaling of ratio in the same way as in the SLIC paper
# so the values have the same meaning
step = float(max((step_z, step_y, step_x)))
ratio = 1.0 / compactness
#我类个去,分割时方不方的骚操作
image = np.ascontiguousarray(image * ratio)
labels = _slic_cython(image, segments, step, max_iter, spacing, slic_zero)
#把过小过小的处理一下
if enforce_connectivity:
segment_size = depth * height * width / n_segments
min_size = int(min_size_factor * segment_size)
max_size = int(max_size_factor * segment_size)
labels = _enforce_label_connectivity_cython(labels,
min_size,
max_size)
if is_2d:
labels = labels[0]
return labels参考:csdn