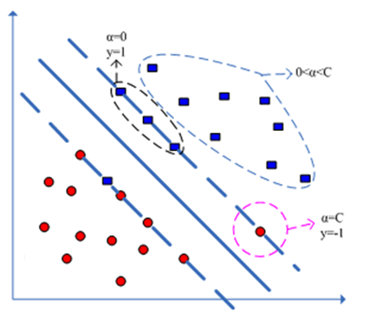
纵观SMO算法,其核心是怎么选择每轮优化的两个拉格朗日乘子,标准的SMO算法是通过判断乘子是否违反原问题的KKT条件来选择待优化乘子的,这里可能有一个问题,回顾原问题的KKT条件:
是否违反它,与这几个因素相关:拉格朗日乘子、样本标记
、偏置
。
的更新依赖于两个优化拉格朗日乘子,这就可能出现这种情况:拉格朗日乘子
已经能使目标函数达到最优,而SMO算法本身并不能确定当前由于两个优化拉格朗日乘子计算得到的
是否就是使目标函数达到最优的那个
,换句话说,对一些本来不违反KKT条件的点,由于上次迭代选择了不合适的
,使得它们出现违反KKT条件的情况,导致后续出现一些耗时而无用的搜索,针对标准SMO的缺点,出现了以下几种改进方法,它们同样是通过KKT条件来选择乘子,不同之处为使用对偶问题的KKT条件。
1、通过Maximal Violating Pair选择优化乘子
原问题的对偶问题指的是:
它的拉格朗日方程可以写为:
定义,则对偶问题的KKT条件为:
总结以下可分为3种情况:
分类标记可以取1或-1,按照正类和负类区分指标集,
、
、
、
、
,如图:
图一
整理一下就得到KKT条件的新形式:
从图上也可以看到,当分类器对全部样本都分类正确的时候,必有:
像标准SMO一样,要精确达到最优值显然也是没必要的,因此这里也需要加个容忍值。
引入容忍值后的KKT条件为:
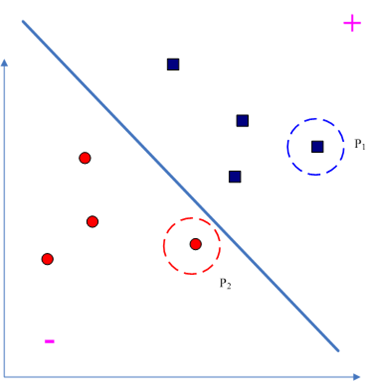
根据前面的说明可以定义“违反对”:
那么我们到底要优化哪两个拉格朗日乘子呢?由“违反对”的定义可知,和
差距最大的那两个点最违反KKT条件的,描述为:
于是就是一个“Maximal Violating Pair,一个示意图如下,其中
和
就是一对MVP:
2、通过Second Order Information选择优化乘子
不论是通过一阶信息还是二阶信息来选择优化乘子,都是基于以下这个定理:
定理一:如果矩阵为半正定矩阵,当且仅当待优化乘子为“违反对”时,对于SMO类型的算法,其目标函数严格递减(如:
)。
First Order Information选择方法我就不介绍了,直接看Second Order Information选择方法:
当选定一个乘子后,另一个乘子选择的条件是使得当前“乘子对”为“违反对”且能使目标函数值最小,在最优化迭代类型方法中非常重要的一个工具就是函数的taylor展开式,优化的过程可以概括为一系列选搜索方向、选搜索步长的过程,对目标函数w展开得:
,这里d分别为两个待选择乘子的优化方向。
于是最小化就变成了:
由优化条件可知:
这样选择乘子的过程就变成了以下优化问题:
总结通过Second Order Information选择优化乘子的方法如下:
1、首先选择一个乘子,条件如下:
2、选择另外一个乘子的过程就是求解的过程,设
为第二个乘子,则有:
将条件:
带入有:
于是目标函数在处达到最小值:
用式子表示就是:
当这种情况出现时可以用一个很小的值来代替它,具体可见《A study on SMO-type decomposition methods for support vector machines》一文中“Non-Positive Definite Kernel Matrices”小节。
4、算法实现
将代码加入到了LeftNotEasy的pymining项目中了,可以从http://code.google.com/p/python-data-mining-platform/下载。
几点说明:
1、训练和测试数据格式为:value1 value2 value3 …… valuen,label=+1,数据集放在***.data中,对应的label放在***.labels中,也可以使用libsvm的数据集;
2、分类器目前支持的核为RBF、Linear、Polynomial、Sigmoid、将来支持String Kernel;
3、训练集和测试集的输入支持dense matrix 和 sparse matrix,其中sparse matrix采用CSR表示法;
4、对于不平衡数据的处理一般来说从三个方面入手:
1)、对正例和负例赋予不同的C值,例如正例远少于负例,则正例的C值取得较大,这种方法的缺点是可能会偏离原始数据的概率分布;
2)、对训练集的数据进行预处理即对数量少的样本以某种策略进行采样,增加其数量或者减少数量多的样本,典型的方法如:随机插入法,缺点是可能出现
overfitting,较好的是:Synthetic Minority Over-sampling TEchnique(SMOTE),其缺点是只能应用在具体的特征空间中,不适合处理那些无法用
特征向量表示的问题,当然增加样本也意味着训练时间可能增加;
3)、基于核函数的不平衡数据处理。
本文就以容易实现为原则,采用第一种方式,配置文件中的节点<c>中的值代表的是正例的代价值,模型中的npRatio用来计算负例的代价值,例如正例远多于负例,则这个比率就大于一,反之亦然,原理如下图所示:
和
异号且
的情形(
的情况类似)
和
同号的情形
5、算法最耗时的地方是优化乘子的选择和更新一阶导数信息,这两个地方都需要去计算核函数值,而核函数值的计算最终都需要去做内积运算,这就意味着原始空间的维度很高会增加内积运算的时间;对于dense matrix我就直接用numpy的dot了,而sparse matrix采用的是CSR表示法,求它的内积我实验过的方法有三种,第一种不需要额外空间,但时间复杂度为O(nlgn),第二种需要一个hash表(用dictionary代替了),时间复杂度为线性,第三种需要一个bitmap(使用BitVector),时间复杂度也为线性,实际使用中第一种速度最快,我就暂时用它了,应该还有更快的方法,希望高人们能指点一下;另外由于使用dictionary缓存核矩阵,遇到训练数据很大的数据集很容易挂掉,所以在程序中,当dictionary的内存占用达到配置文件的阈值时会将其中相对次要的元素删掉,保留对角线上的内积值。
6、使用psyco
相对于c,python在进行高维度数值计算时表现的会比较差,为了加快数值计算的速度,我使用了psyco来进行加速,它的使用很简单,加两句话就行,用程序测试一下使用psyco之前和之后的表现:
定义一个1164维的向量,代码如下:
1: import time
2: import psyco
3: psyco.full()
4:
5: def dot():
6: vec =[0.0081484941037,0.088244064232,-0.0116517274912,0.0175709303652,-0.0626241393835,-0.0854107365903,-0.0351575090354,0.0380456765128,0.0180095979816,-0.0209914241928,0.163106660981,0.127053320056,-0.0335430235607,-0.0109767864904,0.0709995225815,0.0167076809919,0.027726097262,-0.0311204836595,-0.0259199476295,0.180810035412,-0.0075547491507,-0.0081006110325,0.0270585413405,-0.0148313935713,0.0337146203424,0.0736048267004,-0.182810465676,-0.0267899729122,0.000179219280158,-0.0981396318556,0.0162153149149,0.0158205889243,0.0135732439379,-0.0122793913668,-0.0248816557428,-0.0839578376417,0.00599848509353,0.114707262204,-0.0328007287819,0.0467560652885,0.313179556657,0.024121214556,-0.0553332419857,-0.00684296908044,0.174685776575,0.00418298437096,0.015624947957,-0.0357577181683,-0.0335658241296,-0.0413500938049,0.00890996222676,-0.191520167295,-0.02223857119,-0.00900410728578,-0.00101642281143,-0.0751063838566,-0.00779308778792,-0.0339048097301,-0.29873321989,-0.00751397119331,0.0020689071961,-0.0334433996129,0.0798526578962,-0.00314060942661,0.000994142874497,-0.00385208084616,0.00639638012896,0.00432347948574,-0.0295723516279,-0.0282409806726,-0.243877258982,0.0306503469536,-0.0229985748532,0.0284501391965,-0.198174324275,-0.0179711916393,0.0143267555469,-0.0287066071347,0.000282523969584,0.0616495560776,-0.0172220711262,-0.0687083629936,-0.00370051948645,-0.0219797414557,0.15847905705,0.0354483161821,0.0763267553204,-0.0429680349387,0.438909021868,-0.0549235929403,-0.00176280411289,0.0168842161271,-0.0245477451033,0.103534103547,0.190875334952,-0.0160077971357,-1.13916725717,-0.0578280272522,0.618283515386,0.0218745277242,-0.0637461100123,0.00666421528081,0.0276254007423,-0.042759151837,-0.0145850642315,-0.00880446485194,0.000126716038755,0.00433140420099,0.0308245755082,-0.00623921726713,-0.0318217746795,-0.0382440579868,0.000428347914493,0.000530399307534,-0.0759717737756,0.0354515033891,0.130639143673,-0.141494874567,0.0838856126349,-0.0194418010364,0.0137972983237,0.000531338760413,0.00134150184801,0.261957858812,0.0237711884021,0.0230326428759,0.0219937855594,0.00719528755352,0.0333485055281,0.00833840466389,-0.0250022671701,0.0732187999699,0.0409463350505,-0.00163879777058,0.0323944604152,0.0248038687327,0.00763331851835,-0.00540476791599,-0.0700331001035,0.636770876239,0.0270714128914,-0.0562305127792,0.0369742780132,-0.00482469423333,-0.153208622043,-0.169948249631,0.0822114655752,-0.000810202457017,0.0592939745916,0.0210041288368,0.0424686903816,0.013082261434,-0.0270151903807,0.0226204321573,0.00337807861336,0.0552972148331,0.00137329198924,0.00410977518032,-0.0788631223297,0.0195763268983,-0.011867418399,0.000136137516823,0.0489199529798,-0.0272194722771,0.0126117026801,-0.422521768213,0.0175167663074,-0.513577519799,-0.304908016713,-0.0153815043354,0.0143756230195,0.038892601783,0.00785100547614,0.024633644749,0.0565278241742,-0.019980734894,0.100938716186,0.0274989424604,0.0103429343526,-0.0533430239958,0.0319011843986,-0.0168859775771,-0.0443695710743,-0.0079129398118,-0.0125144644331,0.13605025411,-0.0162963376194,-0.000710295461299,0.0144422401202,-0.0184620116687,0.0804442274609,-0.0234468286624,-0.0238108738443,0.00860171509498,0.0081484941037,0.088244064232,-0.0116517274912,0.0175709303652,-0.0626241393835,-0.0854107365903,-0.0351575090354,0.0380456765128,0.0180095979816,-0.0209914241928,0.163106660981,0.127053320056,-0.0335430235607,-0.0109767864904,0.0709995225815,0.0167076809919,0.027726097262,-0.0311204836595,-0.0259199476295,0.180810035412,-0.0075547491507,-0.0081006110325,0.0270585413405,-0.0148313935713,0.0337146203424,0.0736048267004,-0.182810465676,-0.0267899729122,0.000179219280158,-0.0981396318556,0.0162153149149,0.0158205889243,0.0135732439379,-0.0122793913668,-0.0248816557428,-0.0839578376417,0.00599848509353,0.114707262204,-0.0328007287819,0.0467560652885,0.313179556657,0.024121214556,-0.0553332419857,-0.00684296908044,0.174685776575,0.00418298437096,0.015624947957,-0.0357577181683,-0.0335658241296,-0.0413500938049,0.00890996222676,-0.191520167295,-0.02223857119,-0.00900410728578,-0.00101642281143,-0.0751063838566,-0.00779308778792,-0.0339048097301,-0.29873321989,-0.00751397119331,0.0020689071961,-0.0334433996129,0.0798526578962,-0.00314060942661,0.000994142874497,-0.00385208084616,0.00639638012896,0.00432347948574,-0.0295723516279,-0.0282409806726,-0.243877258982,0.0306503469536,-0.0229985748532,0.0284501391965,-0.198174324275,-0.0179711916393,0.0143267555469,-0.0287066071347,0.000282523969584,0.0616495560776,-0.0172220711262,-0.0687083629936,-0.00370051948645,-0.0219797414557,0.15847905705,0.0354483161821,0.0763267553204,-0.0429680349387,0.438909021868,-0.0549235929403,-0.00176280411289,0.0168842161271,-0.0245477451033,0.103534103547,0.190875334952,-0.0160077971357,-1.13916725717,-0.0578280272522,0.618283515386,0.0218745277242,-0.0637461100123,0.00666421528081,0.0276254007423,-0.042759151837,-0.0145850642315,-0.00880446485194,0.000126716038755,0.00433140420099,0.0308245755082,-0.00623921726713,-0.0318217746795,-0.0382440579868,0.000428347914493,0.000530399307534,-0.0759717737756,0.0354515033891,0.130639143673,-0.141494874567,0.0838856126349,-0.0194418010364,0.0137972983237,0.000531338760413,0.00134150184801,0.261957858812,0.0237711884021,0.0230326428759,0.0219937855594,0.00719528755352,0.0333485055281,0.00833840466389,-0.0250022671701,0.0732187999699,0.0409463350505,-0.00163879777058,0.0323944604152,0.0248038687327,0.00763331851835,-0.00540476791599,-0.0700331001035,0.636770876239,0.0270714128914,-0.0562305127792,0.0369742780132,-0.00482469423333,-0.153208622043,-0.169948249631,0.0822114655752,-0.000810202457017,0.0592939745916,0.0210041288368,0.0424686903816,0.013082261434,-0.0270151903807,0.0226204321573,0.00337807861336,0.0552972148331,0.00137329198924,0.00410977518032,-0.0788631223297,0.0195763268983,-0.011867418399,0.000136137516823,0.0489199529798,-0.0272194722771,0.0126117026801,-0.422521768213,0.0175167663074,-0.513577519799,-0.304908016713,-0.0153815043354,0.0143756230195,0.038892601783,0.00785100547614,0.024633644749,0.0565278241742,-0.019980734894,0.100938716186,0.0274989424604,0.0103429343526,-0.0533430239958,0.0319011843986,-0.0168859775771,-0.0443695710743,-0.0079129398118,-0.0125144644331,0.13605025411,-0.0162963376194,-0.000710295461299,0.0144422401202,-0.0184620116687,0.0804442274609,-0.0234468286624,-0.0238108738443,0.00860171509498,0.0081484941037,0.088244064232,-0.0116517274912,0.0175709303652,-0.0626241393835,-0.0854107365903,-0.0351575090354,0.0380456765128,0.0180095979816,-0.0209914241928,0.163106660981,0.127053320056,-0.0335430235607,-0.0109767864904,0.0709995225815,0.0167076809919,0.027726097262,-0.0311204836595,-0.0259199476295,0.180810035412,-0.0075547491507,-0.0081006110325,0.0270585413405,-0.0148313935713,0.0337146203424,0.0736048267004,-0.182810465676,-0.0267899729122,0.000179219280158,-0.0981396318556,0.0162153149149,0.0158205889243,0.0135732439379,-0.0122793913668,-0.0248816557428,-0.0839578376417,0.00599848509353,0.114707262204,-0.0328007287819,0.0467560652885,0.313179556657,0.024121214556,-0.0553332419857,-0.00684296908044,0.174685776575,0.00418298437096,0.015624947957,-0.0357577181683,-0.0335658241296,-0.0413500938049,0.00890996222676,-0.191520167295,-0.02223857119,-0.00900410728578,-0.00101642281143,-0.0751063838566,-0.00779308778792,-0.0339048097301,-0.29873321989,-0.00751397119331,0.0020689071961,-0.0334433996129,0.0798526578962,-0.00314060942661,0.000994142874497,-0.00385208084616,0.00639638012896,0.00432347948574,-0.0295723516279,-0.0282409806726,-0.243877258982,0.0306503469536,-0.0229985748532,0.0284501391965,-0.198174324275,-0.0179711916393,0.0143267555469,-0.0287066071347,0.000282523969584,0.0616495560776,-0.0172220711262,-0.0687083629936,-0.00370051948645,-0.0219797414557,0.15847905705,0.0354483161821,0.0763267553204,-0.0429680349387,0.438909021868,-0.0549235929403,-0.00176280411289,0.0168842161271,-0.0245477451033,0.103534103547,0.190875334952,-0.0160077971357,-1.13916725717,-0.0578280272522,0.618283515386,0.0218745277242,-0.0637461100123,0.00666421528081,0.0276254007423,-0.042759151837,-0.0145850642315,-0.00880446485194,0.000126716038755,0.00433140420099,0.0308245755082,-0.00623921726713,-0.0318217746795,-0.0382440579868,0.000428347914493,0.000530399307534,-0.0759717737756,0.0354515033891,0.130639143673,-0.141494874567,0.0838856126349,-0.0194418010364,0.0137972983237,0.000531338760413,0.00134150184801,0.261957858812,0.0237711884021,0.0230326428759,0.0219937855594,0.00719528755352,0.0333485055281,0.00833840466389,-0.0250022671701,0.0732187999699,0.0409463350505,-0.00163879777058,0.0323944604152,0.0248038687327,0.00763331851835,-0.00540476791599,-0.0700331001035,0.636770876239,0.0270714128914,-0.0562305127792,0.0369742780132,-0.00482469423333,-0.153208622043,-0.169948249631,0.0822114655752,-0.000810202457017,0.0592939745916,0.0210041288368,0.0424686903816,0.013082261434,-0.0270151903807,0.0226204321573,0.00337807861336,0.0552972148331,0.00137329198924,0.00410977518032,-0.0788631223297,0.0195763268983,-0.011867418399,0.000136137516823,0.0489199529798,-0.0272194722771,0.0126117026801,-0.422521768213,0.0175167663074,-0.513577519799,-0.304908016713,-0.0153815043354,0.0143756230195,0.038892601783,0.00785100547614,0.024633644749,0.0565278241742,-0.019980734894,0.100938716186,0.0274989424604,0.0103429343526,-0.0533430239958,0.0319011843986,-0.0168859775771,-0.0443695710743,-0.0079129398118,-0.0125144644331,0.13605025411,-0.0162963376194,-0.000710295461299,0.0144422401202,-0.0184620116687,0.0804442274609,-0.0234468286624,-0.0238108738443,0.00860171509498,0.0081484941037,0.088244064232,-0.0116517274912,0.0175709303652,-0.0626241393835,-0.0854107365903,-0.0351575090354,0.0380456765128,0.0180095979816,-0.0209914241928,0.163106660981,0.127053320056,-0.0335430235607,-0.0109767864904,0.0709995225815,0.0167076809919,0.027726097262,-0.0311204836595,-0.0259199476295,0.180810035412,-0.0075547491507,-0.0081006110325,0.0270585413405,-0.0148313935713,0.0337146203424,0.0736048267004,-0.182810465676,-0.0267899729122,0.000179219280158,-0.0981396318556,0.0162153149149,0.0158205889243,0.0135732439379,-0.0122793913668,-0.0248816557428,-0.0839578376417,0.00599848509353,0.114707262204,-0.0328007287819,0.0467560652885,0.313179556657,0.024121214556,-0.0553332419857,-0.00684296908044,0.174685776575,0.00418298437096,0.015624947957,-0.0357577181683,-0.0335658241296,-0.0413500938049,0.00890996222676,-0.191520167295,-0.02223857119,-0.00900410728578,-0.00101642281143,-0.0751063838566,-0.00779308778792,-0.0339048097301,-0.29873321989,-0.00751397119331,0.0020689071961,-0.0334433996129,0.0798526578962,-0.00314060942661,0.000994142874497,-0.00385208084616,0.00639638012896,0.00432347948574,-0.0295723516279,-0.0282409806726,-0.243877258982,0.0306503469536,-0.0229985748532,0.0284501391965,-0.198174324275,-0.0179711916393,0.0143267555469,-0.0287066071347,0.000282523969584,0.0616495560776,-0.0172220711262,-0.0687083629936,-0.00370051948645,-0.0219797414557,0.15847905705,0.0354483161821,0.0763267553204,-0.0429680349387,0.438909021868,-0.0549235929403,-0.00176280411289,0.0168842161271,-0.0245477451033,0.103534103547,0.190875334952,-0.0160077971357,-1.13916725717,-0.0578280272522,0.618283515386,0.0218745277242,-0.0637461100123,0.00666421528081,0.0276254007423,-0.042759151837,-0.0145850642315,-0.00880446485194,0.000126716038755,0.00433140420099,0.0308245755082,-0.00623921726713,-0.0318217746795,-0.0382440579868,0.000428347914493,0.000530399307534,-0.0759717737756,0.0354515033891,0.130639143673,-0.141494874567,0.0838856126349,-0.0194418010364,0.0137972983237,0.000531338760413,0.00134150184801,0.261957858812,0.0237711884021,0.0230326428759,0.0219937855594,0.00719528755352,0.0333485055281,0.00833840466389,-0.0250022671701,0.0732187999699,0.0409463350505,-0.00163879777058,0.0323944604152,0.0248038687327,0.00763331851835,-0.00540476791599,-0.0700331001035,0.636770876239,0.0270714128914,-0.0562305127792,0.0369742780132,-0.00482469423333,-0.153208622043,-0.169948249631,0.0822114655752,-0.000810202457017,0.0592939745916,0.0210041288368,0.0424686903816,0.013082261434,-0.0270151903807,0.0226204321573,0.00337807861336,0.0552972148331,0.00137329198924,0.00410977518032,-0.0788631223297,0.0195763268983,-0.011867418399,0.000136137516823,0.0489199529798,-0.0272194722771,0.0126117026801,-0.422521768213,0.0175167663074,-0.513577519799,-0.304908016713,-0.0153815043354,0.0143756230195,0.038892601783,0.00785100547614,0.024633644749,0.0565278241742,-0.019980734894,0.100938716186,0.0274989424604,0.0103429343526,-0.0533430239958,0.0319011843986,-0.0168859775771,-0.0443695710743,-0.0079129398118,-0.0125144644331,0.13605025411,-0.0162963376194,-0.000710295461299,0.0144422401202,-0.0184620116687,0.0804442274609,-0.0234468286624,-0.0238108738443,0.00860171509498,0.0081484941037,0.088244064232,-0.0116517274912,0.0175709303652,-0.0626241393835,-0.0854107365903,-0.0351575090354,0.0380456765128,0.0180095979816,-0.0209914241928,0.163106660981,0.127053320056,-0.0335430235607,-0.0109767864904,0.0709995225815,0.0167076809919,0.027726097262,-0.0311204836595,-0.0259199476295,0.180810035412,-0.0075547491507,-0.0081006110325,0.0270585413405,-0.0148313935713,0.0337146203424,0.0736048267004,-0.182810465676,-0.0267899729122,0.000179219280158,-0.0981396318556,0.0162153149149,0.0158205889243,0.0135732439379,-0.0122793913668,-0.0248816557428,-0.0839578376417,0.00599848509353,0.114707262204,-0.0328007287819,0.0467560652885,0.313179556657,0.024121214556,-0.0553332419857,-0.00684296908044,0.174685776575,0.00418298437096,0.015624947957,-0.0357577181683,-0.0335658241296,-0.0413500938049,0.00890996222676,-0.191520167295,-0.02223857119,-0.00900410728578,-0.00101642281143,-0.0751063838566,-0.00779308778792,-0.0339048097301,-0.29873321989,-0.00751397119331,0.0020689071961,-0.0334433996129,0.0798526578962,-0.00314060942661,0.000994142874497,-0.00385208084616,0.00639638012896,0.00432347948574,-0.0295723516279,-0.0282409806726,-0.243877258982,0.0306503469536,-0.0229985748532,0.0284501391965,-0.198174324275,-0.0179711916393,0.0143267555469,-0.0287066071347,0.000282523969584,0.0616495560776,-0.0172220711262,-0.0687083629936,-0.00370051948645,-0.0219797414557,0.15847905705,0.0354483161821,0.0763267553204,-0.0429680349387,0.438909021868,-0.0549235929403,-0.00176280411289,0.0168842161271,-0.0245477451033,0.103534103547,0.190875334952,-0.0160077971357,-1.13916725717,-0.0578280272522,0.618283515386,0.0218745277242,-0.0637461100123,0.00666421528081,0.0276254007423,-0.042759151837,-0.0145850642315,-0.00880446485194,0.000126716038755,0.00433140420099,0.0308245755082,-0.00623921726713,-0.0318217746795,-0.0382440579868,0.000428347914493,0.000530399307534,-0.0759717737756,0.0354515033891,0.130639143673,-0.141494874567,0.0838856126349,-0.0194418010364,0.0137972983237,0.000531338760413,0.00134150184801,0.261957858812,0.0237711884021,0.0230326428759,0.0219937855594,0.00719528755352,0.0333485055281,0.00833840466389,-0.0250022671701,0.0732187999699,0.0409463350505,-0.00163879777058,0.0323944604152,0.0248038687327,0.00763331851835,-0.00540476791599,-0.0700331001035,0.636770876239,0.0270714128914,-0.0562305127792,0.0369742780132,-0.00482469423333,-0.153208622043,-0.169948249631,0.0822114655752,-0.000810202457017,0.0592939745916,0.0210041288368,0.0424686903816,0.013082261434,-0.0270151903807,0.0226204321573,0.00337807861336,0.0552972148331,0.00137329198924,0.00410977518032,-0.0788631223297,0.0195763268983,-0.011867418399,0.000136137516823,0.0489199529798,-0.0272194722771,0.0126117026801,-0.422521768213,0.0175167663074,-0.513577519799,-0.304908016713,-0.0153815043354,0.0143756230195,0.038892601783,0.00785100547614,0.024633644749,0.0565278241742,-0.019980734894,0.100938716186,0.0274989424604,0.0103429343526,-0.0533430239958,0.0319011843986,-0.0168859775771,-0.0443695710743,-0.0079129398118,-0.0125144644331,0.13605025411,-0.0162963376194,-0.000710295461299,0.0144422401202,-0.0184620116687,0.0804442274609,-0.0234468286624,-0.0238108738443,0.00860171509498,0.0081484941037,0.088244064232,-0.0116517274912,0.0175709303652,-0.0626241393835,-0.0854107365903,-0.0351575090354,0.0380456765128,0.0180095979816,-0.0209914241928,0.163106660981,0.127053320056,-0.0335430235607,-0.0109767864904,0.0709995225815,0.0167076809919,0.027726097262,-0.0311204836595,-0.0259199476295,0.180810035412,-0.0075547491507,-0.0081006110325,0.0270585413405,-0.0148313935713,0.0337146203424,0.0736048267004,-0.182810465676,-0.0267899729122,0.000179219280158,-0.0981396318556,0.0162153149149,0.0158205889243,0.0135732439379,-0.0122793913668,-0.0248816557428,-0.0839578376417,0.00599848509353,0.114707262204,-0.0328007287819,0.0467560652885,0.313179556657,0.024121214556,-0.0553332419857,-0.00684296908044,0.174685776575,0.00418298437096,0.015624947957,-0.0357577181683,-0.0335658241296,-0.0413500938049,0.00890996222676,-0.191520167295,-0.02223857119,-0.00900410728578,-0.00101642281143,-0.0751063838566,-0.00779308778792,-0.0339048097301,-0.29873321989,-0.00751397119331,0.0020689071961,-0.0334433996129,0.0798526578962,-0.00314060942661,0.000994142874497,-0.00385208084616,0.00639638012896,0.00432347948574,-0.0295723516279,-0.0282409806726,-0.243877258982,0.0306503469536,-0.0229985748532,0.0284501391965,-0.198174324275,-0.0179711916393,0.0143267555469,-0.0287066071347,0.000282523969584,0.0616495560776,-0.0172220711262,-0.0687083629936,-0.00370051948645,-0.0219797414557,0.15847905705,0.0354483161821,0.0763267553204,-0.0429680349387,0.438909021868,-0.0549235929403,-0.00176280411289,0.0168842161271,-0.0245477451033,0.103534103547,0.190875334952,-0.0160077971357,-1.13916725717,-0.0578280272522,0.618283515386,0.0218745277242,-0.0637461100123,0.00666421528081,0.0276254007423,-0.042759151837,-0.0145850642315,-0.00880446485194,0.000126716038755,0.00433140420099,0.0308245755082,-0.00623921726713,-0.0318217746795,-0.0382440579868,0.000428347914493,0.000530399307534,-0.0759717737756,0.0354515033891,0.130639143673,-0.141494874567,0.0838856126349,-0.0194418010364,0.0137972983237,0.000531338760413,0.00134150184801,0.261957858812,0.0237711884021,0.0230326428759,0.0219937855594,0.00719528755352,0.0333485055281,0.00833840466389,-0.0250022671701,0.0732187999699,0.0409463350505,-0.00163879777058,0.0323944604152,0.0248038687327,0.00763331851835,-0.00540476791599,-0.0700331001035,0.636770876239,0.0270714128914,-0.0562305127792,0.0369742780132,-0.00482469423333,-0.153208622043,-0.169948249631,0.0822114655752,-0.000810202457017,0.0592939745916,0.0210041288368,0.0424686903816,0.013082261434,-0.0270151903807,0.0226204321573,0.00337807861336,0.0552972148331,0.00137329198924,0.00410977518032,-0.0788631223297,0.0195763268983,-0.011867418399,0.000136137516823,0.0489199529798,-0.0272194722771,0.0126117026801,-0.422521768213,0.0175167663074,-0.513577519799,-0.304908016713,-0.0153815043354,0.0143756230195,0.038892601783,0.00785100547614,0.024633644749,0.0565278241742,-0.019980734894,0.100938716186,0.0274989424604,0.0103429343526,-0.0533430239958,0.0319011843986,-0.0168859775771,-0.0443695710743,-0.0079129398118,-0.0125144644331,0.13605025411,-0.0162963376194,-0.000710295461299,0.0144422401202,-0.0184620116687,0.0804442274609,-0.0234468286624,-0.0238108738443,0.00860171509498]
7: sum = 0.0
8: for i in range(len(vec)):
9: sum += vec[i] * vec[i]
10: return sum
11:
12: if __name__ == "__main__":
13: start = time.clock()
14:
15: for i in range(100000):
16: dot()
17:
18: print '\n dot product = ',dot(),'\n'
19:
20: end = time.clock()
21: print end - start, 'seconds.'
注释掉psyco.full()这句话后执行时间为:26.48s,去掉注释后执行时间为:4.6s,改成C代码执行时间为:0.58s,一般情况下psyco都会加快运行速度,当然和C相比其实还是差距明显的,相关链接如下:
1)、可爱的 Python: 用 Psyco 让 Python 运行得像 C 一样快》:http://www.ibm.com/developerworks/cn/linux/sdk/python/charm-28/
2)、psyco主页:http://psyco.sourceforge.net/download.html,在ubuntu下可以这么安装:sudo apt-get install python-psyco
7、关于分类器的评价采用如下指标:
1)、
2)、
3)、
4)、
5)、
6)、
7)、、
8、运行环境:
OS:32bits ubuntu 10.04
CPU:Intel(R) Pentium(R) Dual CPU E2200 @ 2.20GHz
memory:DIMM DDR2 Synchronous 667 MHz (1.5 ns) 2G
IDE:Eclipse + Pydev
9、程序中用到的数据集主要来自UCI Data Set和Libsvm Data:
1)、pymining data:data/train.txt和data/test.txt
DATA ------- 0 ex. ------- 1 ex. -------- 2 ex. ---------- 3 ex. ------------ TotalTraining set 149 --------- 36 --------- 102 ---------- 63 ------------ 350
Validation set 85 --------- 14 --------- 35 ---------- 16 ------------ 150
Number of features:卡方检验过滤后
Total:1862
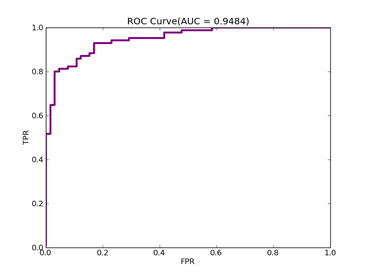
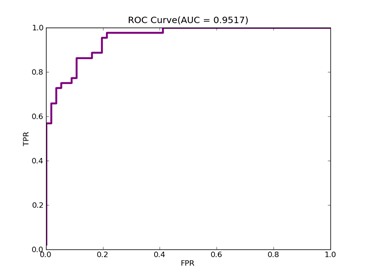
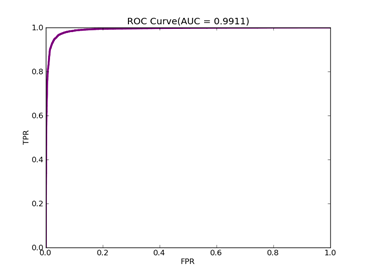
C = 100,kernel = RBF,gamma = 0.03,npRatio=1,测试结果如下:
(1)、类别0标记为1,其它标记为-1,
Recall = 0.929411764706 Precision = 0.840425531915 Accuracy = 0.86
F(beta=1) = 0.564005241516 F(beta=2) = 0.676883364786 AUCb = 0.849321266968
(2)、类别1标记为1,其它标记为-1,
Recall = 0.929411764706 Precision = 0.840425531915 Accuracy = 0.86
F(beta=1) = 0.564005241516 F(beta=2) = 0.676883364786 AUCb = 0.849321266968
(3)、类别2标记为1,其它标记为-1,
Recall = 0.6 Precision = 0.913043478261 Accuracy = 0.893333333333
F(beta=1) = 0.43598615917 F(beta=2) = 0.496845425868 AUCb = 0.791304347826
(4)、类别3标记为1,其它标记为-1,
Recall = 0.25 Precision = 1.0 Accuracy = 0.92
F(beta=1) = 0.222222222222 F(beta=2) = 0.238095238095 AUCb = 0.625
2)、Arcene:http://archive.ics.uci.edu/ml/datasets/Arcene
ARCENE -- Positive ex. -- Negative ex. – Total
Training set 44 ----------- 56 --------- 100
Validation set 44 ----------- 56 --------- 100
Number of variables/features/attributes:
Real: 7000
Probes: 3000
Total: 10000
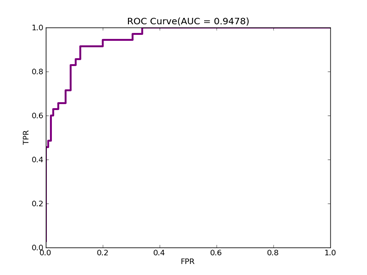
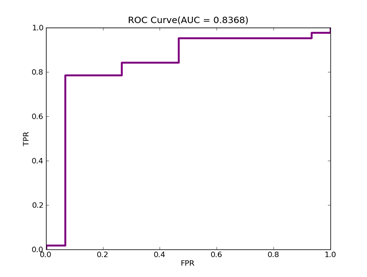
C = 100,kernel = RBF,gamma = 0.000000001,npRatio=1,测试结果如下:
Recall = 0.772727272727 Precision = 0.85 Accuracy = 0.84
F(beta=1) = 0.500866551127 F(beta=2) = 0.584074373484 AUCb = 0.832792207792
3)、SPECT Heart Data Set :http://archive.ics.uci.edu/ml/datasets/SPECT+Heart
SPECT ------ Positive ex. -- Negative ex. – Total
Training set 40------------ 40---------- 80
Validation set 172 ---------- 15--------- 187
Attribute Information:
OVERALL_DIAGNOSIS: 0,1 (class attribute, binary)
F1: 0,1 (the partial diagnosis 1, binary)
F2: 0,1 (the partial diagnosis 2, binary)
F3: 0,1 (the partial diagnosis 3, binary)
F4: 0,1 (the partial diagnosis 4, binary)
F5: 0,1 (the partial diagnosis 5, binary)
F6: 0,1 (the partial diagnosis 6, binary)
F7: 0,1 (the partial diagnosis 7, binary)
F8: 0,1 (the partial diagnosis 8, binary)
F9: 0,1 (the partial diagnosis 9, binary)
F10: 0,1 (the partial diagnosis 10, binary)
F11: 0,1 (the partial diagnosis 11, binary)
F12: 0,1 (the partial diagnosis 12, binary)
F13: 0,1 (the partial diagnosis 13, binary)
F14: 0,1 (the partial diagnosis 14, binary)
F15: 0,1 (the partial diagnosis 15, binary)
F16: 0,1 (the partial diagnosis 16, binary)
F17: 0,1 (the partial diagnosis 17, binary)
F18: 0,1 (the partial diagnosis 18, binary)
F19: 0,1 (the partial diagnosis 19, binary)
F20: 0,1 (the partial diagnosis 20, binary)
F21: 0,1 (the partial diagnosis 21, binary)
F22: 0,1 (the partial diagnosis 22, binary)
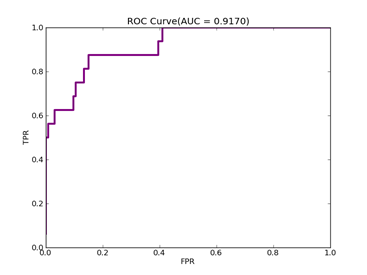
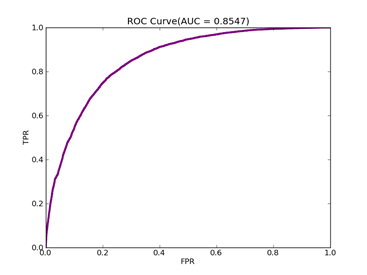
C = 100,kernel = RBF,gamma = 15,npRatio=1,测试结果如下:
Recall = 0.93023255814 Precision = 0.958083832335 Accuracy = 0.898395721925
F(beta=1) = 0.617135142954 F(beta=2) = 0.756787437329 AUCb = 0.731782945736
4)、Dexter:http://archive.ics.uci.edu/ml/datasets/Dexter
DEXTER -- Positive ex. -- Negative ex. – Total
Training set 150 ---------- 150 -------- 300
Validation set 150 ---------- 150 -------- 300
Test set 1000 --------- 1000 ------- 2000 (缺少label)
Number of variables/features/attributes:
Real: 9947
Probes: 10053
Total: 20000
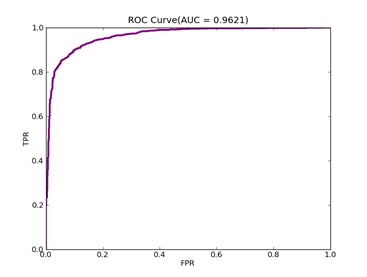
C = 100,kernel = RBF, gamma = 0.000001,npRatio=1,测试结果如下:
Recall = 0.986666666667 Precision = 0.822222222222 Accuracy = 0.886666666667
F(beta=1) = 0.577637130802 F(beta=2) = 0.698291252232 AUCb = 0.886666666667
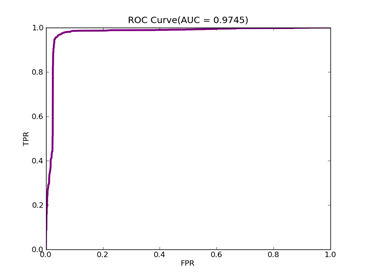
 5)、Mushrooms:原始数据集在:http://archive.ics.uci.edu/ml/datasets/Mushroom
5)、Mushrooms:原始数据集在:http://archive.ics.uci.edu/ml/datasets/Mushroom
预处理以后的在:http://www.csie.ntu.edu.tw/~cjlin/libsvmtools/datasets/binary/mushrooms
MUSHROOMS-- Positive ex.(lbael=1) -- Negative ex.(lbael=2) – Total
Data set 3916 ----------------------- 4208 ------------------- 8124
C = 100,kernel = RBF,gamma = 0.00001,npRatio=1,测试结果如下:
Recall = 1.0 Precision = 0.942615239887 Accuracy = 0.960897435897
F(beta=1) = 0.640664961637 F(beta=2) = 0.793097989552 AUCb = 0.945340501792
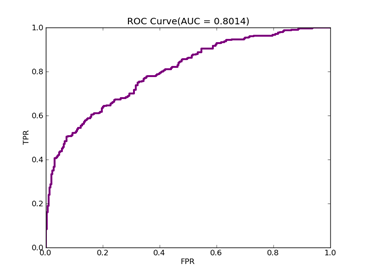
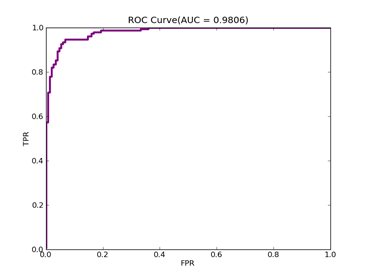
 6)、 Adult:原始数据集在:http://archive.ics.uci.edu/ml/datasets/Adult
6)、 Adult:原始数据集在:http://archive.ics.uci.edu/ml/datasets/Adult
预处理以后的在:http://www.csie.ntu.edu.tw/~cjlin/libsvmtools/datasets/binary.html a1a
ADULT ------ Positive ex.(lbael=+1) --- Negative ex.(lbael=-1) – Total
Training set 395 -------------------- 1210 ----------------- 1605
Validation set 7446 -------------------- 23510 ----------------- 30956
Number of variables/features/attributes: 123 C = 100,kernel = RBF,gamma = 0.08,npRatio=1,测试结果如下:
Recall = 0.598710717164 Precision = 0.588281868567 Accuracy = 0.802687685748
F(beta=1) = 0.322095887953 F(beta=2) = 0.339513363093 AUCb = 0.733000615919
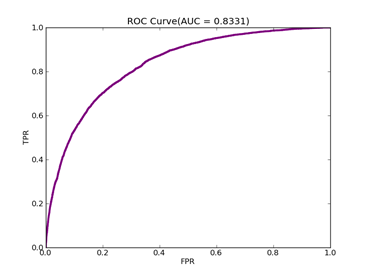 由于正例少于负例,调整模型的npRatio=10,测试结果如下:
由于正例少于负例,调整模型的npRatio=10,测试结果如下:
Recall = 0.620198764437 Precision = 0.602243088159 Accuracy = 0.810117586251
F(beta=1) = 0.336126156669 F(beta=2) = 0.357601319181 AUCb = 0.745233367757
其它参数不变,npRatio=100,测试结果如下:
Recall = 0.6952726296 Precision = 0.595399654974 Accuracy = 0.813057242538
F(beta=1) = 0.361435449815 F(beta=2) = 0.39122162696 AUCb = 0.772817088939
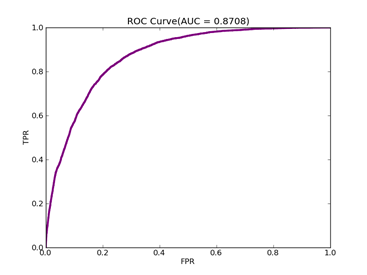 可以从上面的实验看到处理不平衡数据时候,对正例和负例赋予不同的C值方法的作用。
可以从上面的实验看到处理不平衡数据时候,对正例和负例赋予不同的C值方法的作用。
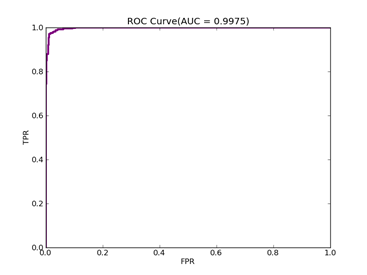
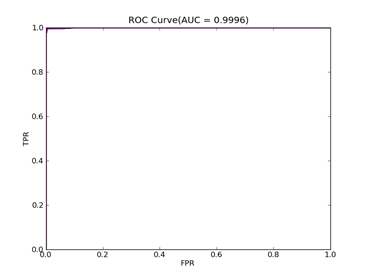
7)、Fourclass: 数据集在:http://www.csie.ntu.edu.tw/~cjlin/libsvmtools/datasets/binary/fourclass
FOURCLASS-- Positive ex.(lbael=1) -- Negative ex.(lbael=2) – Total
Fourclass set 307 ----------------------- 557 ------------------- 864
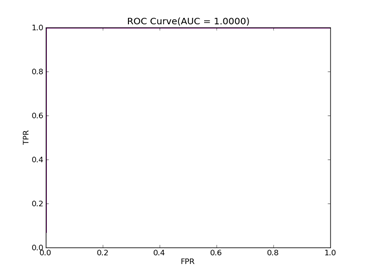
C = 100,kernel = RBF,gamma = 0.01,npRatio=1,测试结果如下:
Recall = 0.991071428571 Precision = 1.0 Accuracy = 0.996415770609
F(beta=1) = 0.662686567164 F(beta=2) = 0.827123695976 AUCb = 0.995535714286
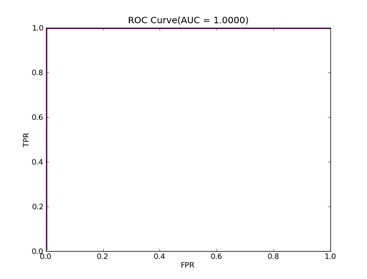 8)、Splice:http://www.csie.ntu.edu.tw/~cjlin/libsvmtools/datasets/binary.html splice
8)、Splice:http://www.csie.ntu.edu.tw/~cjlin/libsvmtools/datasets/binary.html splice
SPLICE-- Positive ex.(lbael=1) -- Negative ex.(lbael=2) – Total
Training set 517 --------------------- 483 --------------- 1000
Validation set 1131 -------------------- 1044 -------------- 2175
Number of variables/features/attributes: 60 C = 100,kernel = RBF,gamma = 0.01,npRatio=1,测试结果如下:
Recall = 0.885057471264 Precision = 0.912488605287 Accuracy = 0.896091954023
F(beta=1) = 0.577366617352 F(beta=2) = 0.696505768897 AUCb = 0.896551724138
9)、RCV1.BINARY:http://www.csie.ntu.edu.tw/~cjlin/libsvmtools/datasets/binary.html
RCV1.BINARY------Total
Training set 20242
Validation set 677399
Number of features:
Total: 47236
Recall = 0.956610366919 Precision = 0.964097045588 Accuracy = 0.9593
F(beta=1) = 0.631535514017 F(beta=2) = 0.778847158177 AUCb = 0.959383756361
10)、Madelon:http://archive.ics.uci.edu/ml/datasets/Madelon
MADELON -- Positive ex. -- Negative ex. – Total
Training set 1000 --------- 1000 ------- 2000
Validation set 300 ---------- 300 -------- 600
Test set 900 ---------- 900 -------- 1800
Number of variables/features/attributes:
Real: 20
Probes: 480
Total: 500
Recall = 0.673333333333 Precision = 0.724014336918 Accuracy = 0.708333333333
F(beta=1) = 0.406701950583 F(beta=2) = 0.451613474471 AUCb = 0.708333333333
11)、GISETTE:http://archive.ics.uci.edu/ml/datasets/Gisette
GISETTE -- Positive ex. -- Negative ex. – Total
Training set 3000 -------- 3000 ------- 6000
Validation set 500 --------- 500 ---------1000
Test set 3250 -------- 3250 ------- 6500
Number of variables/features/attributes:
Real: 2500
Probes: 2500
Total: 5000
Recall = 0.972 Precision = 0.983805668016 Accuracy = 0.978
F(beta=1) = 0.647037875094 F(beta=2) = 0.802795761493 AUCb = 0.978
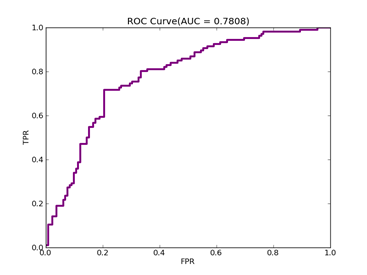
12)、Australian:http://www.csie.ntu.edu.tw/~cjlin/libsvmtools/datasets/binary.html
GISETTE -- Positive ex. -- Negative ex. – Total
Training set 201 --------- 251 -------- 452
Validation set 106 --------- 132 ---------238
Number of features:
Total: 14
Recall = 0.801886792453 Precision = 0.643939393939 Accuracy = 0.714285714286
F(beta=1) = 0.422243001578 F(beta=2) = 0.474093808236 AUCb = 0.722913093196
13)、Svmguide1:http://www.csie.ntu.edu.tw/~cjlin/libsvmtools/datasets/binary.html
GISETTE -- Positive ex. -- Negative ex. – Total
Training set 2000 --------- 1089 -------- 3089
Validation set 2000 --------- 2000 ---------4000
Number of features:
Total: 4
Recall = 0.964 Precision = 0.957774465971 Accuracy = 0.96075
F(beta=1) = 0.632009483243 F(beta=2) = 0.7795759451 AUCb = 0.96075
5、并行点
如果期望在并行计算框架中运行SMO,我可以想到的并行点如下:
1)、选择最大违反对时,可以先在每台机器上找到局部MVP,最后由master选择全局MVP;
2)、更新一阶导数数组,各个机器更新自己本地部分就可以了;
3)、对sparse matrix向量求内积,可以用map函数映射为key/value对,其中key为column标号,value为对应的取值,reduce函数根据key计算乘积并累加。
想要使两个拉格朗日乘子的优化过程实现并行似乎很有难度,目前没有想到好方法。
6、总结
本文介绍了如何通过优化拉格朗日乘子的选择来提高SMO算法性能的方法,SMO算法的核心思想是每次选择两个拉格朗日乘子进行优化,因此其主要矛盾就落在了优先优化哪些乘子身上,目前对乘子选择最好的方法之一是利用对偶问题的KKT条件来寻找最大违反对(MVP),而寻找MVP的方法又可以利用一阶信息和二阶信息,其中又以后者效果较好,它对MVP的选定不仅考虑它们之间值的差距,而且考虑两个乘子之间的伪距离
。
如果数据分布不平衡,训练出的分类器精度等常规指标可能很高,但是实际应用时就会出现问题,典型的例子是漏油检测,处理这类问题可以从三个方面入手:预处理数据集本身、使用不同惩罚系数、使用特殊核函数,它们各有优缺点,没有最好只有最合适,本文采用第二种方法,实现起来也比较容易,评价指标采用ROC Curve、AUC,本文绘制ROC的方法采用Libsvm中的方法。
参数选择直接影响到分类器的性能,涉及的主要参数有:惩罚系数C,这个值越大代表越不想放弃离群点;npRatio,如果数据分布很不平衡,可以用它来调节惩罚系数,负例远大于正例时,npRatio>1,负例的C值要小于正例的C值,反之亦然;核函数参数,依据具体核函数确定。本文没有给出选择最优参数的方法。
7、参考文献
1)、《Improvements to Platt’s SMO Algorithm for SVM Classifier Design》
2)、《A Study on SMO-type Decomposition Methods for Support Vector Machines》
3)、《The Analysis of Decomposition Methods for Support Vector Machines》
4)、《Working Set Selection Using Second Order Information for Training Support Vector Machines》
5)、《Making Large-Scale SVM Learning Practical》
6)、《Optimizing Area Under Roc Curve with SVMs》
7)、《Applying Support Vector Machines to Imbalanced Datasets》
8)、《SVMs Modeling for Highly Imbalanced》
转载于:https://www.cnblogs.com/vivounicorn/archive/2011/08/25/2152824.html